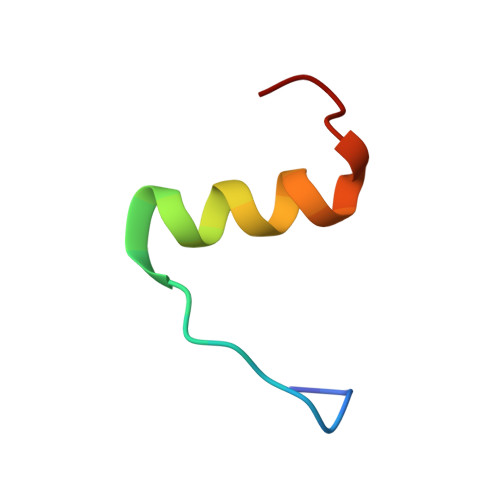
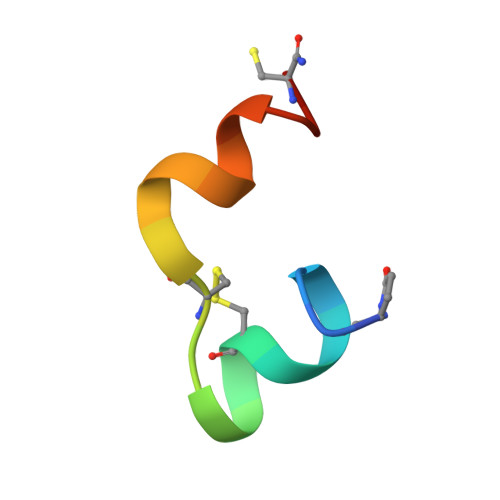Structure of the R3/I5 Chimeric Relaxin Peptide, a Selective GPCR135 and GPCR142 Agonist.
Haugaard-Jonsson, L.M., Hossain, M.A., Daly, N.L., Bathgate, R.A., Wade, J.D., Craik, D.J., Rosengren, K.J.(2008) J Biological Chem 283: 23811-23818
- PubMed: 18577524
- DOI: https://doi.org/10.1074/jbc.M800489200
- Primary Citation of Related Structures:
2K1V - PubMed Abstract:
The human relaxin family comprises seven peptide hormones with various biological functions mediated through interactions with G-protein-coupled receptors. Interestingly, among the hitherto characterized receptors there is no absolute selectivity toward their primary ligand. The most striking example of this is the relaxin family ancestor, relaxin-3, which is an agonist for three of the four currently known relaxin receptors: GPCR135, GPCR142, and LGR7. Relaxin-3 and its endogenous receptor GPCR135 are both expressed predominantly in the brain and have been linked to regulation of stress and feeding. However, to fully understand the role of relaxin-3 in neurological signaling, the development of selective GPCR135 agonists and antagonists for in vivo studies is crucial. Recent reports have demonstrated that such selective ligands can be achieved by making chimeric peptides comprising the relaxin-3 B-chain combined with the INSL5 A-chain. To obtain structural insights into the consequences of combining A- and B-chains from different relaxins we have determined the NMR solution structure of a human relaxin-3/INSL5 chimeric peptide. The structure reveals that the INSL5 A-chain adopts a conformation similar to the relaxin-3 A-chain, and thus has the ability to structurally support a native-like conformation of the relaxin-3 B-chain. These findings suggest that the decrease in activity at the LGR7 receptor seen for this peptide is a result of the removal of a secondary LGR7 binding site present in the relaxin-3 A-chain, rather than conformational changes in the primary B-chain receptor binding site.
- School of Pure and Applied Natural Sciences, University of Kalmar, SE-391 82 Kalmar, Sweden.
Organizational Affiliation: