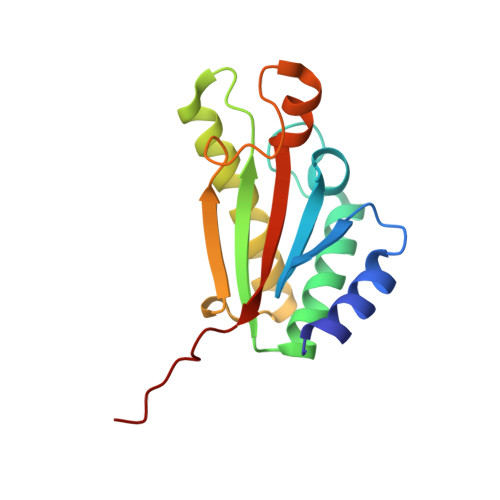
Solution NMR structure of the folded C-terminal fragment of YiaD from Escherichia coli. Northeast Structural Genomics Consortium target ER553.
Ramelot, T.A., Zhao, L., Hamilton, K., Maglaqui, M., Xiao, R., Liu, J., Baran, M.C., Swapna, G., Acton, T.B., Rost, B., Montelione, G.T., Kennedy, M.A.To be published.