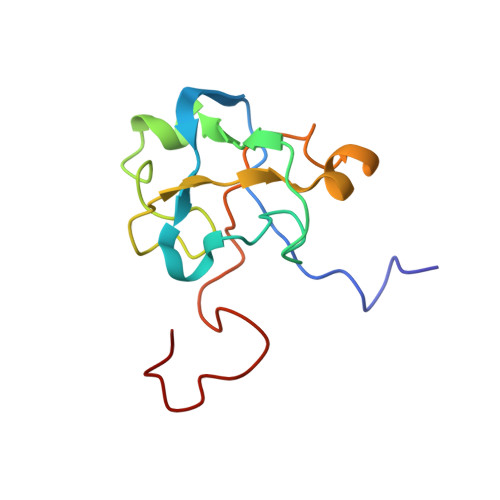
Solution structure of the U2 snRNP protein Rds3p reveals a knotted zinc-finger motif.
van Roon, A.M., Loening, N.M., Obayashi, E., Yang, J.C., Newman, A.J., Hernandez, H., Nagai, K., Neuhaus, D.(2008) Proc Natl Acad Sci U S A 105: 9621-9626
- PubMed: 18621724
- DOI: https://doi.org/10.1073/pnas.0802494105
- Primary Citation of Related Structures:
2K0A - PubMed Abstract:
Rds3p, a component of the U2 snRNP subcomplex SF3b, is essential for pre-mRNA splicing and is extremely well conserved in all eukaryotic species. We report here the solution structure of Rds3p, which reveals an unusual knotted fold unrelated to previously known knotted proteins. Rds3p has a triangular shape with a GATA-like zinc finger at each vertex. Pairs of cysteines contributing to each finger are arranged nonsequentially in a permuted arrangement reminiscent of domain-swapping but which here involves segments of subdomains within a single chain. We suggest that the structure arose through a process of segment swapping after gene duplication. The fingers are connected through beta-strands and loops, forming an overall topology strongly resembling a "triquetra knot." The conservation and surface properties of Rds3p suggest that it functions as a platform for protein assembly within the multiprotein SF3b complex of U2 snRNP. The recombinant protein used for structure determination is biologically active, as it restores splicing activity in a yeast splicing extract depleted of native Rds3p.
- Medical Research Council Laboratory of Molecular Biology, Hills Road, Cambridge, CB2 0QH, United Kingdom.
Organizational Affiliation: