structural basis of transcription elongation control: the NusA-aCTD complex
Prasch, S., Schweimer, K., Roesch, P.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA-directed RNA polymerase subunit alpha | 99 | Yersinia pseudotuberculosis | Mutation(s): 0 Gene Names: rpoA EC: 2.7.7.6 | 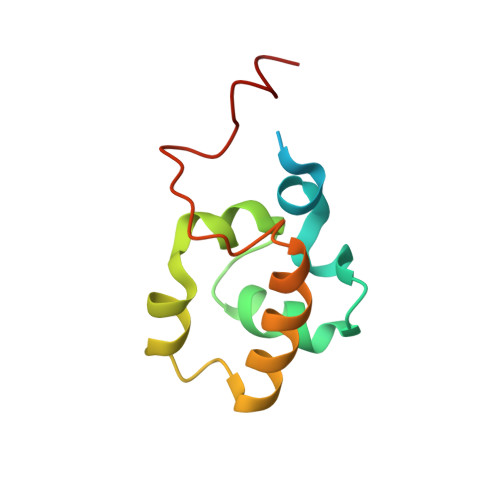 | |
UniProt | |||||
Find proteins for Q664U6 (Yersinia pseudotuberculosis serotype I (strain IP32953)) Explore Q664U6 Go to UniProtKB: Q664U6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q664U6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Transcription elongation protein nusA | 74 | Escherichia coli | Mutation(s): 0 Gene Names: nusA | 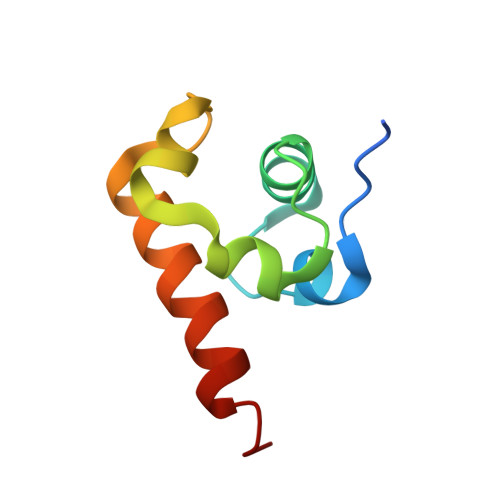 | |
UniProt | |||||
Find proteins for P0AFF6 (Escherichia coli (strain K12)) Explore P0AFF6 Go to UniProtKB: P0AFF6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0AFF6 | ||||
Sequence AnnotationsExpand | |||||
| |||||