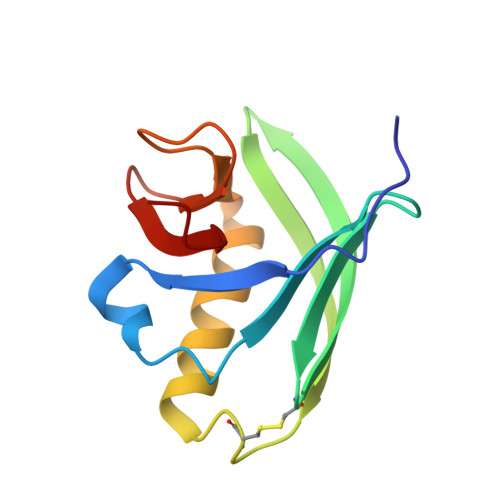
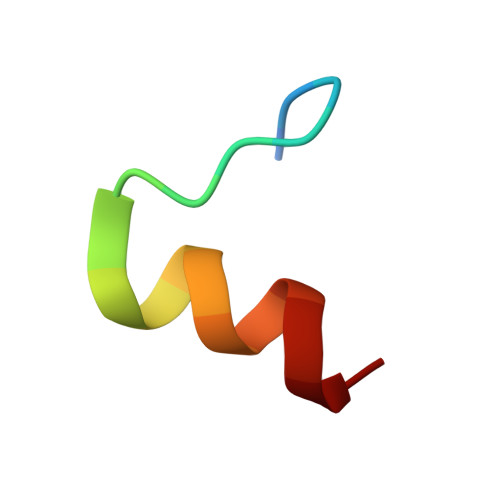
Structural Characterization of the Type-III Pilot-Secretin Complex from Shigella flexneri
Okon, M., Moraes, T.F., Lario, P.I., Creagh, A.L., Haynes, C.A., Strynadka, N.C., McIntosh, L.P.(2008) Structure 16: 1544-1554
- PubMed: 18940609
- DOI: https://doi.org/10.1016/j.str.2008.08.006
- Primary Citation of Related Structures:
2JW1 - PubMed Abstract:
Assembly of the type-III secretion apparatus, which translocates proteins through both membranes of Gram-negative bacterial pathogens into host cells, requires the formation of an integral outer-membrane secretin ring. Typically, a small lipidated pilot protein is necessary for the stabilization and localization of this ring. Using NMR spectroscopy, we demonstrate that the C-terminal residues 553-570 of the Shigella flexneri secretin MxiD encompass the minimal binding domain for its cognate pilot MxiM. Although unstructured in isolation, upon complex formation with MxiM, these residues fold into an amphipathic turn-helix motif that caps the elongated hydrophobic cavity of the cracked beta-barrel pilot. Along with a rearrangement of core aromatic residues, this prevents the binding of lipids within the cavity. The mutually exclusive association of lipids and MxiD with MxiM establishes a framework for understanding the role of a pilot in the outer-membrane insertion and multimerization of the secretin ring.
- Department of Biochemistry & Molecular Biology, University of British Columbia, Vancouver, BC V6T 1Z3, Canada.
Organizational Affiliation: