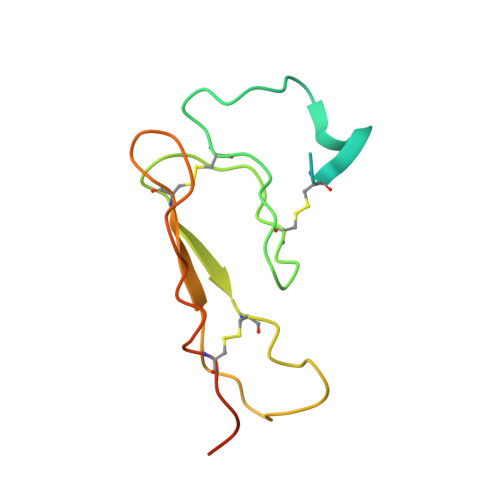
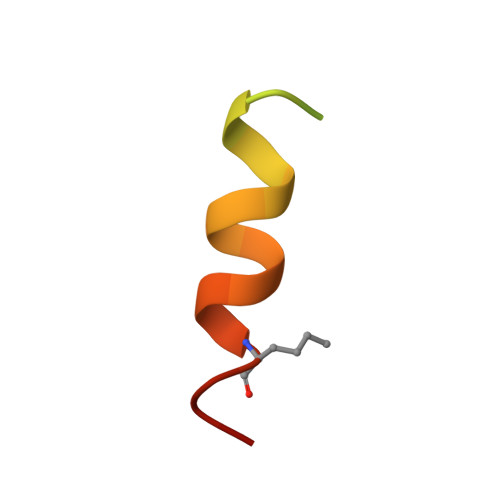
Structure of the N-terminal domain of a type B1 G protein-coupled receptor in complex with a peptide ligand
Grace, C.R.R., Perrin, M.H., Jozsef, G., DiGruccio, M.R., Cantle, J.P., Rivier, J.E., Vale, W.W., Riek, R.(2007) Proc Natl Acad Sci U S A 104: 4858-4863
- PubMed: 17360332
- DOI: https://doi.org/10.1073/pnas.0700682104
- Primary Citation of Related Structures:
2JNC, 2JND - PubMed Abstract:
The corticotropin releasing factor (CRF) family of ligands and their receptors coordinate endocrine, behavioral, autonomic, and metabolic responses to stress and play additional roles within the cardiovascular, gastrointestinal, and other systems. The actions of CRF and the related urocortins are mediated by activation of two receptors, CRF-R1 and CRF-R2, belonging to the B1 family of G protein-coupled receptors. The short-consensus-repeat fold (SCR) within the first extracellular domain (ECD1) of the CRF receptor(s) comprises the major ligand binding site and serves to dock a peptide ligand via its C-terminal segment, thus positioning the N-terminal segment to interact with the receptor's juxtamembrane domains to activate the receptor. Here we present the 3D NMR structure of ECD1 of CRF-R2beta in complex with astressin, a peptide antagonist. In the structure of the complex the C-terminal segment of astressin forms an amphipathic helix, whose entire hydrophobic face interacts with the short-consensus-repeat motif, covering a large intermolecular interface. In addition, the complex is characterized by intermolecular hydrogen bonds and a salt bridge. These interactions are quantitatively weighted by an analysis of the effects on the full-length receptor affinities using an Ala scan of CRF. These structural studies identify the major determinants for CRF ligand specificity and selectivity and support a two-step model for receptor activation. Furthermore, because of a proposed conservation of the fold for both the ECD1s and ligands, this structure can serve as a model for ligand recognition for the entire B1 receptor family.
- The Clayton Foundation Laboratories for Peptide Biology, The Salk Institute for Biological Studies, 10010 North Torrey Pines Road, La Jolla, CA 92037, USA.
Organizational Affiliation: