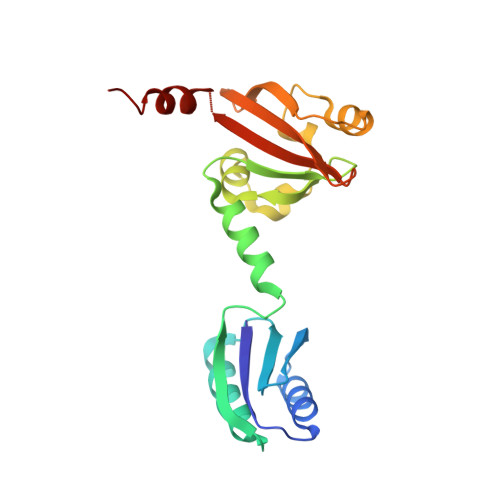
Crystal Structure of the N-Terminal Domain of the Tyrr Transcription Factor Responsible for Gene Regulation of Aromatic Amino Acid Biosynthesis and Transport in Escherichia Coli K12
Verger, D., Carr, P.D., Kwok, T., Ollis, D.L.(2007) J Mol Biology 367: 102
- PubMed: 17222426
- DOI: https://doi.org/10.1016/j.jmb.2006.12.018
- Primary Citation of Related Structures:
2JHE - PubMed Abstract:
The X-ray structure of the N-terminal domain of TyrR has been solved to a resolution of 2.3 A. It reveals a modular protein containing an ACT domain, a connecting helix, a PAS domain and a C-terminal helix. Two dimers are present in the asymmetric unit with one monomer of each pair exhibiting a large rigid-body movement that results in a hinging around residue 74 of approximately 50 degrees . The structure of the dimer is discussed with reference to other transcription regulator proteins. Putative binding sites are identified for the aromatic amino acid cofactors.
- School of Crystallography, Birkbeck College, University of London, UK.
Organizational Affiliation: