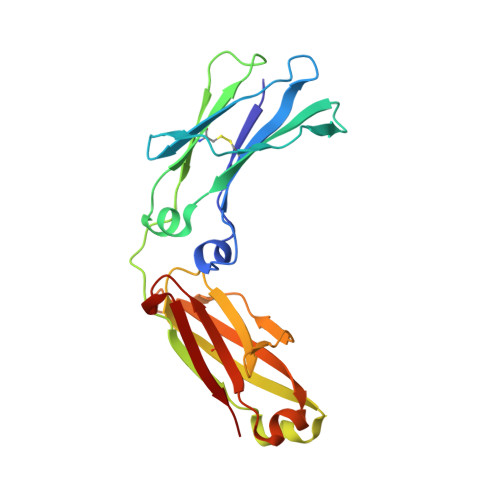
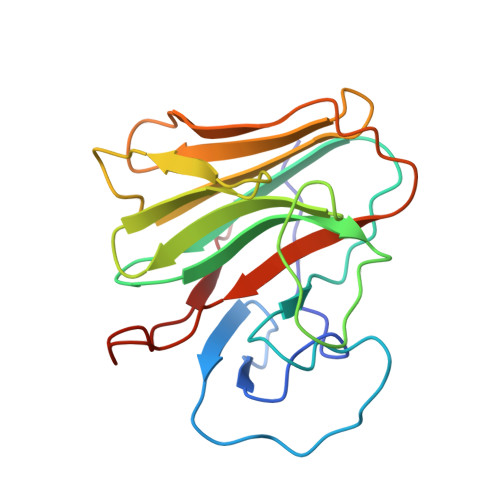
Structural Basis for Pryspry-Mediated Tripartite Motif (Trim) Protein Function.
James, L.C., Keeble, A.H., Khan, Z., Rhodes, D.A., Trowsdale, J.(2007) Proc Natl Acad Sci U S A 104: 6200
- PubMed: 17400754
- DOI: https://doi.org/10.1073/pnas.0609174104
- Primary Citation of Related Structures:
2IWG - PubMed Abstract:
The human tripartite motif (TRIM) family comprises 70 members, including HIV restriction factor TRIM5alpha and disease-associated proteins TRIM20 (pyrin) and TRIM21. TRIM proteins have conserved domain architecture but diverse cellular roles. Here, we describe how the C-terminal PRYSPRY domain mediates diverse TRIM functions. The crystal structure of TRIM21 PRYSPRY in complex with its target IgG Fc reveals a canonical binding interface comprised of two discrete pockets formed by antibody-like variable loops. Alanine scanning of this interface has identified the hot-spot residues that control TRIM21 binding to Fc; the same hot-spots control HIV/murine leukemia virus restriction by TRIM5alpha and mediate severe familial Mediterranean fever in TRIM20/pyrin. Characterization of the IgG binding site for TRIM21 PRYSPRY reveals TRIM21 as a superantigen analogous to bacterial protein A and suggests that an antibody bipolar bridging mechanism may contribute to the pathogenic accumulation of anti-TRIM21 autoantibody immune complex in autoimmune disease.
- Protein and Nucleic Acid Chemistry Division, Medical Research Council Laboratory of Molecular Biology, Cambridge CB2 2QH, UK. lcj@mrc-lmb.cam.ac.uk
Organizational Affiliation: