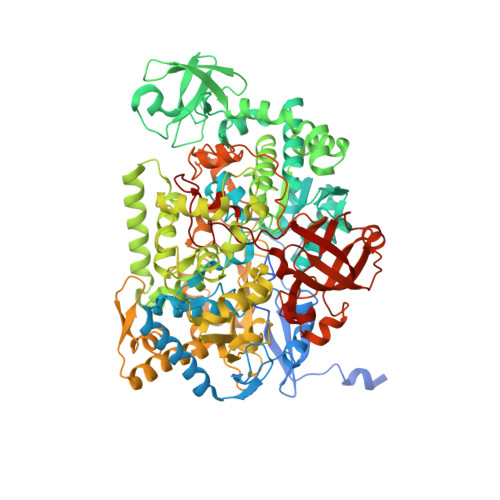
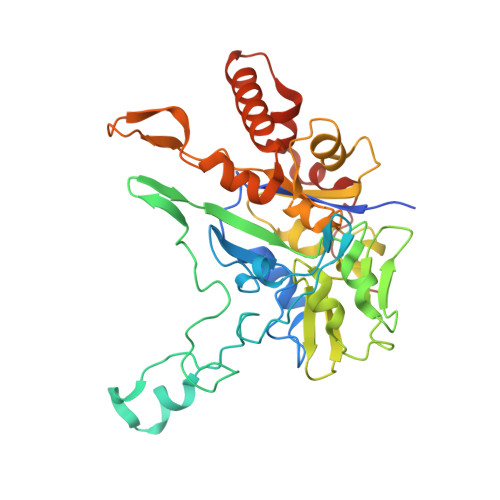
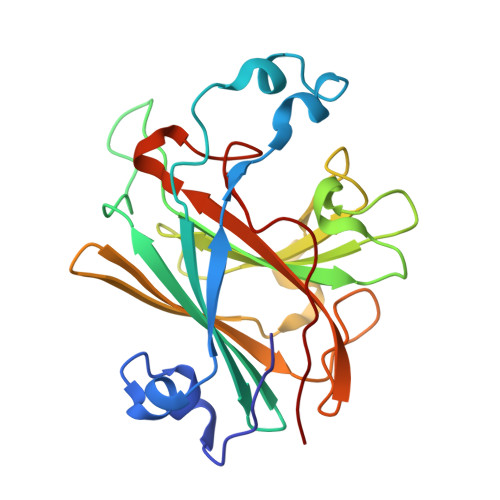
Crystal Structure of Ethylbenzene Dehydrogenase from Aromatoleum Aromaticum
Kloer, D.P., Hagel, C., Heider, J., Schulz, G.E.(2006) Structure 14: 1377
- PubMed: 16962969
- DOI: https://doi.org/10.1016/j.str.2006.07.001
- Primary Citation of Related Structures:
2IVF - PubMed Abstract:
Anaerobic degradation of hydrocarbons was discovered a decade ago, and ethylbenzene dehydrogenase was one of the first characterized enzymes involved. The structure of the soluble periplasmic 165 kDa enzyme was established at 1.88 A resolution. It is a heterotrimer. The alpha subunit contains the catalytic center with a molybdenum held by two molybdopterin-guanine dinucleotides, one with an open pyran ring, and an iron-sulfur cluster with a histidine ligand. During catalysis, electrons produced by substrate oxidation are transferred to a heme in the gamma subunit and then presumably to a separate cytochrome involved in nitrate respiration. The beta subunit contains four iron-sulfur clusters and is structurally related to ferredoxins. The gamma subunit is the first known protein with a methionine and a lysine as axial heme ligands. The catalytic product was modeled into the active center, showing the reaction geometry. A mechanism consistent with activity and inhibition data of ethylbenzene-related compounds is proposed.
- Institut für Organische Chemie und Biochemie, Albert-Ludwigs-Universität, Albertstr. 21, D-79104 Freiburg im Breisgau, Germany.
Organizational Affiliation: