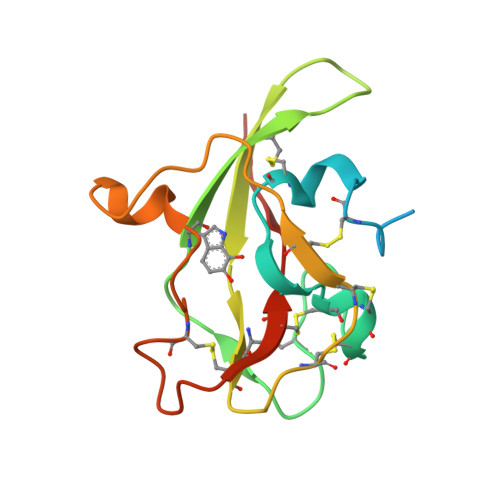
Atomic Level Insight Into the Oxidative Half-Reaction of Aromatic Amine Dehydrogenase.
Roujeinikova, A., Scrutton, N., Leys, D.(2006) J Biological Chem 281: 40264
- PubMed: 17005560
- DOI: https://doi.org/10.1074/jbc.M605559200
- Primary Citation of Related Structures:
2HXC, 2IUP, 2IUQ, 2IUR, 2IUV - PubMed Abstract:
The quinoprotein aromatic amine dehydrogenase (AADH) uses a covalently bound tryptophan tryptophylquinone (TTQ) cofactor to oxidatively deaminate primary aromatic amines. Recent crystal structures have provided insight into the reductive half-reaction. In contrast, no atomic details are available for the oxidative half-reaction. The TTQ O7 hydroxyl group is protonated during reduction, but it is unclear how this proton can be removed during the oxidative half-reaction. Furthermore, compared with the electron transfer from the N-quinol form, electron transfer from the non-physiological O-quinol form to azurin is significantly slower. Here we report crystal structures of the O-quinol, N-quinol, and N-semiquinone forms of AADH. A comparison of oxidized and substrate reduced AADH species reveals changes in the TTQ-containing subunit, extending from residues in the immediate vicinity of the N-quinol to the putative azurin docking site, suggesting a mechanism whereby TTQ redox state influences interprotein electron transfer. In contrast, chemical reduction of the TTQ center has no significant effect on protein conformation. Furthermore, structural reorganization upon substrate reduction places a water molecule near TTQ O7 where it can act as proton acceptor. The structure of the N-semiquinone, however, is essentially similar to oxidized AADH. Surprisingly, in the presence of substrate a covalent N-semiquinone substrate adduct is observed. To our knowledge this is the first detailed insight into a complex, branching mechanism of quinone oxidation where significant structural reorganization upon reduction of the quinone center directly influences formation of the electron transfer complex and nature of the electron transfer process.
- Manchester Interdisciplinary Biocentre, University of Manchester, Manchester M1 7DN, United Kingdom.
Organizational Affiliation: