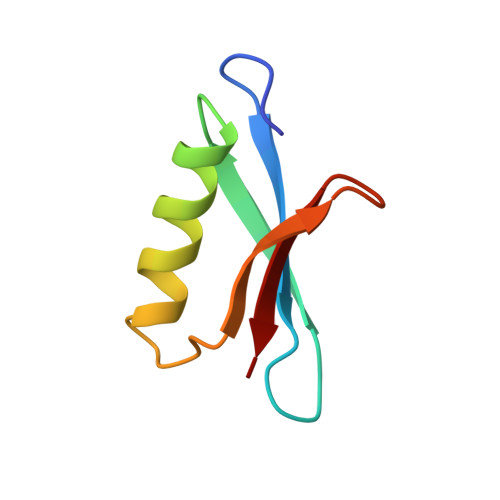
Determination of the solution structures of domains II and III of protein G from Streptococcus by 1H nuclear magnetic resonance.
Lian, L.Y., Derrick, J.P., Sutcliffe, M.J., Yang, J.C., Roberts, G.C.(1992) J Mol Biology 228: 1219-1234
- PubMed: 1474588
- DOI: https://doi.org/10.1016/0022-2836(92)90328-h
- Primary Citation of Related Structures:
2IGG, 2IGH - PubMed Abstract:
We have used 1H nuclear magnetic resonance spectroscopy to determine the solution structures of two small (61 and 64 residue) immunoglobulin G (IgG)-binding domains from protein G, a cell-surface protein from Streptococcus strain G148. The two domains differ in sequence by four amino acid substitutions, and differ in their affinity for some subclasses of IgG. The structure of domain II was determined using a total of 478 distance restraints, 31 phi and 9 chi 1 dihedral angle restraints; that of domain III was determined using a total of 445 distance restraints, 31 phi and 9 chi 1 dihedral angle restraints. A protocol which involved distance geometry, simulated annealing and restrained molecular dynamics was used to determine ensembles of 40 structures consistent with these restraints. The structures are found to consist of an alpha-helix packed against a four-stranded antiparallel-parallel-antiparallel beta-sheet. The structures of the two domains are compared to each other and to the reported structure of a similar domain from a protein G from a different strain of Streptococcus. We conclude that the difference in affinity of domains II and III for IgG is due to local changes in amino acid side-chains, rather than a more extensive change in conformation, suggesting that one or more of the residues which differ between them are directly involved in interaction with IgG.
- Biological N M R Centre, University of Leicester, U.K.
Organizational Affiliation: