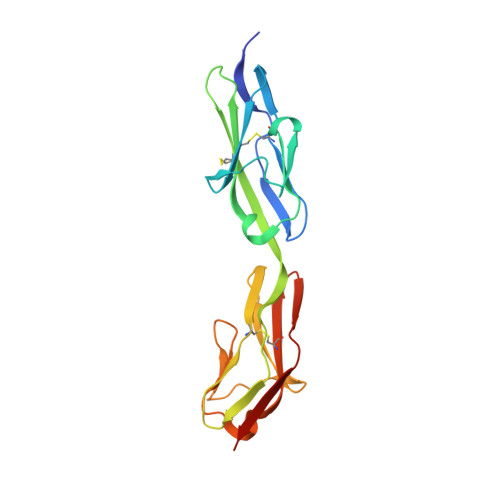
Crystal Structure of the Agrin-responsive Immunoglobulin-like Domains 1 and 2 of the Receptor Tyrosine Kinase MuSK
Stiegler, A.L., Burden, S.J., Hubbard, S.R.(2006) J Mol Biology 364: 424-433
- PubMed: 17011580
- DOI: https://doi.org/10.1016/j.jmb.2006.09.019
- Primary Citation of Related Structures:
2IEP - PubMed Abstract:
Muscle-specific kinase (MuSK) is a receptor tyrosine kinase expressed exclusively in skeletal muscle, where it is required for formation of the neuromuscular junction. MuSK is activated by agrin, a neuron-derived heparan sulfate proteoglycan. Here, we report the crystal structure of the agrin-responsive first and second immunoglobulin-like domains (Ig1 and Ig2) of the MuSK ectodomain at 2.2 A resolution. The structure reveals that MuSK Ig1 and Ig2 are Ig-like domains of the I-set subfamily, which are configured in a linear, semi-rigid arrangement. In addition to the canonical internal disulfide bridge, Ig1 contains a second, solvent-exposed disulfide bridge, which our biochemical data indicate is critical for proper folding of Ig1 and processing of MuSK. Two Ig1-2 molecules form a non-crystallographic dimer that is mediated by a unique hydrophobic patch on the surface of Ig1. Biochemical analyses of MuSK mutants introduced into MuSK(-/-) myotubes demonstrate that residues in this hydrophobic patch are critical for agrin-induced MuSK activation.
- Structural Biology Program, Skirball Institute of Biomolecular Medicine and Department of Pharmacology, New York University School of Medicine, New York, NY 10016, USA.
Organizational Affiliation: