Crystal Structure of uncharacterized conserved archael protein from Methanopyrus kandleri
Bonanno, J.B., Ramagopal, U.A., Dickey, M., Bain, K.T., Powell, A., Ozyurt, S., Wasserman, S., Sauder, J.M., Burley, S.K., Almo, S.C.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Uncharacterized protein conserved in archaea | 131 | Methanopyrus kandleri | Mutation(s): 0 Gene Names: Q8TX89_METKA, MK0786 EC: 4.1.2.25 | 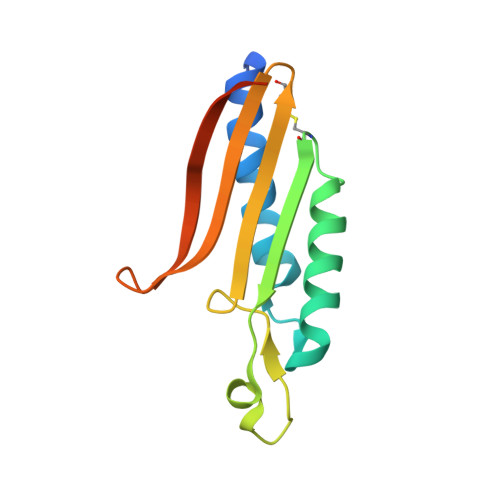 | |
UniProt | |||||
Find proteins for Q8TX89 (Methanopyrus kandleri (strain AV19 / DSM 6324 / JCM 9639 / NBRC 100938)) Explore Q8TX89 Go to UniProtKB: Q8TX89 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8TX89 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| MG Query on MG | E [auth B], F [auth D] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 60.833 | α = 90 |
| b = 60.833 | β = 90 |
| c = 263.945 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| MAR345 | data collection |
| DENZO | data reduction |
| HKL-2000 | data scaling |
| AMoRE | phasing |