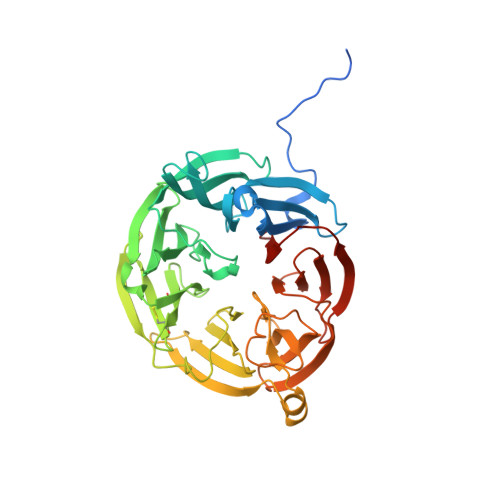
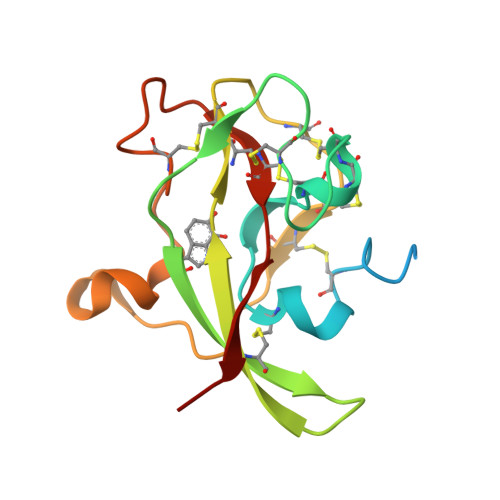
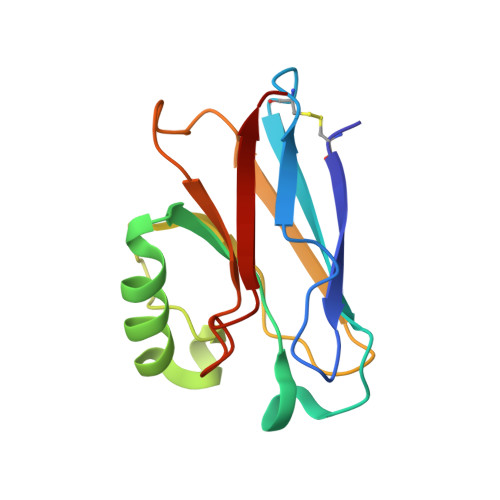
Crystal Structure of an Electron Transfer Complex between Aromatic Amine Dehydrogenase and Azurin from Alcaligenes faecalis.
Sukumar, N., Chen, Z., Ferrari, D., Merli, A., Rossi, G.L., Bellamy, H.D., Chistoserdov, A., Davidson, V.L., Mathews, F.S.(2006) Biochemistry 45: 13500-13510
- PubMed: 17087503
- DOI: https://doi.org/10.1021/bi0612972
- Primary Citation of Related Structures:
2H3X, 2H47, 2IAA - PubMed Abstract:
The crystal structure of an electron transfer complex of aromatic amine dehydrogenase (AADH) and azurin is presented. Electrons are transferred from the tryptophan tryptophylquinone (TTQ) cofactor of AADH to the type I copper of the cupredoxin azurin. This structure is compared with the complex of the TTQ-containing methylamine dehydrogenase (MADH) and the cupredoxin amicyanin. Despite significant similarities between the two quinoproteins and the two cupredoxins, each is specific for its respective partner and the ionic strength dependence and magnitude of the binding constant for each complex are quite different. The AADH-azurin interface is largely hydrophobic, covering approximately 500 A(2) of surface on each molecule, with one direct hydrogen bond linking them. The closest distance from TTQ to copper is 12.6 A compared with a distance of 9.3 A in the MADH-amicyanin complex. When the MADH-amicyanin complex is aligned with the AADH-azurin complex, the amicyanin lies on top of the azurin but is oriented quite differently. Although the copper atoms differ in position by approximately 4.7 A, the amicyanin bound to MADH appears to be rotated approximately 90 degrees from its aligned position with azurin. Comparison of the structures of the two complexes identifies features of the interface that dictate the specificity of the protein-protein interaction and determine the rate of interprotein electron transfer.
- Department of Biochemistry and Molecular Biophysics, Washington University School of Medicine, St. Louis, Missouri 65211, USA.
Organizational Affiliation: