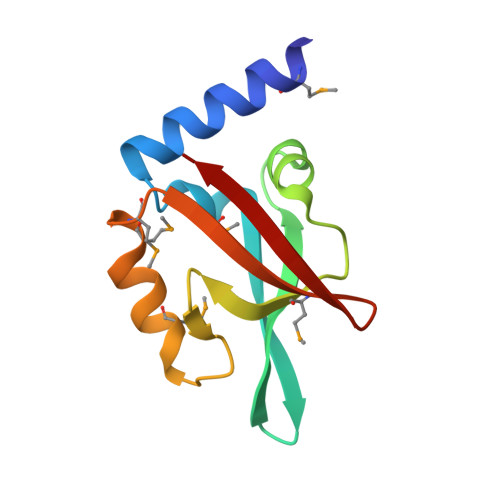
Structural and Functional Studies of EpsC, a Crucial Component of the Type 2 Secretion System from Vibrio cholerae.
Korotkov, K.V., Krumm, B., Bagdasarian, M., Hol, W.G.(2006) J Mol Biology 363: 311-321
- PubMed: 16978643
- DOI: https://doi.org/10.1016/j.jmb.2006.08.037
- Primary Citation of Related Structures:
2I4S, 2I6V - PubMed Abstract:
The type 2 secretion system (T2SS) occurring in Gram-negative bacteria is composed of 12-15 different proteins which form large assemblies spanning two membranes and secreting several virulence factors in folded state across the outer membrane. The T2SS component EpsC of Vibrio cholerae plays an important role in this machinery. While anchored in the inner membrane, by far the largest part of EpsC is periplasmic, containing a so-called homology region (HR) domain and a PDZ domain. Here we report studies on the structure and function of both periplasmic domains of EpsC. The crystal structures of two variants of the PDZ domain of EpsC from V. cholerae were determined at better than 2 A resolution. Compared to the short variant, the longer variant contains an additional N-terminal helix, and reveals a significant difference in the position of helix alphaB with respect to the beta-sheet. Both our structures show that the PDZ domain of EpsC adopts a more open form than in previously reported structures of other PDZ domains. Most interestingly, in the crystals of the short EpsC-PDZ domain the peptide binding groove interacts with an alpha-helix from a neighboring subunit burying approximately 921 A2 solvent accessible surface. This makes it possible that the PDZ domain of this bacterial protein binds proteins in a manner which is altogether different from that seen in any other PDZ domain so far. We also determined that the HR domain of EpsC is primarily responsible for the interaction with the secretin EpsD, while the PDZ is not, or much less, so. This new finding, together with studies of others, leads to the suggestion that the PDZ domain of EpsC may interact with exoproteins to be secreted while the HR domain plays a key role in linking the inner-membrane sub-complex of the T2SS in V. cholerae to the outer membrane secretin.
- Department of Biochemistry, Biomolecular Structure Center, University of Washington, Box 357742, Seattle, WA 98195, USA.
Organizational Affiliation: