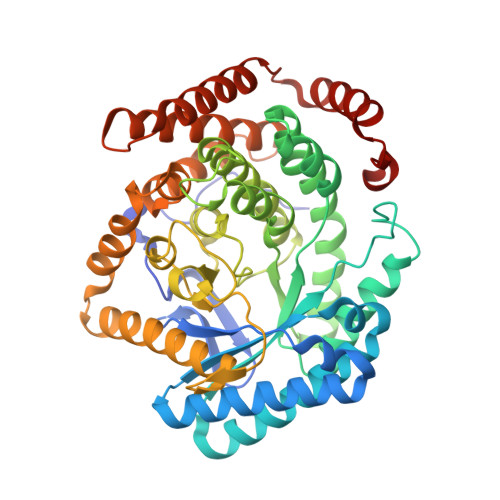
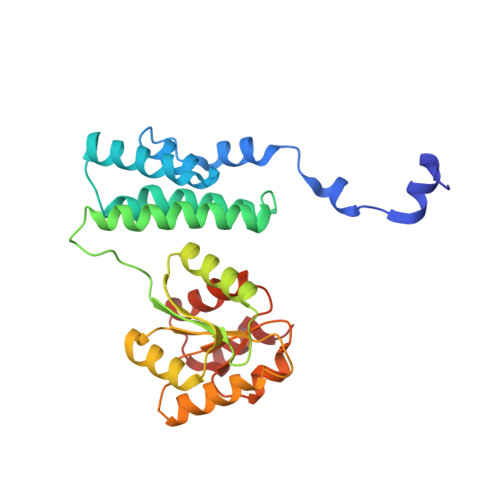
Insight into the mechanism of biological methanol activation based on the crystal structure of the methanol-cobalamin methyltransferase complex
Hagemeier, C.H., Krer, M., Thauer, R.K., Warkentin, E., Ermler, U.(2006) Proc Natl Acad Sci U S A 103: 18917-18922
- PubMed: 17142327
- DOI: https://doi.org/10.1073/pnas.0603650103
- Primary Citation of Related Structures:
2I2X - PubMed Abstract:
Some methanogenic and acetogenic microorganisms have the catalytic capability to cleave heterolytically the C O bond of methanol. To obtain insight into the elusive enzymatic mechanism of this challenging chemical reaction we have investigated the methanol-activating MtaBC complex from Methanosarcina barkeri composed of the zinc-containing MtaB and the 5-hydroxybenzimidazolylcobamide-carrying MtaC subunits. Here we report the 2.5-A crystal structure of this complex organized as a (MtaBC)(2) heterotetramer. MtaB folds as a TIM barrel and contains a novel zinc-binding motif. Zinc(II) lies at the bottom of a funnel formed at the C-terminal beta-barrel end and ligates to two cysteinyl sulfurs (Cys-220 and Cys-269) and one carboxylate oxygen (Glu-164). MtaC is structurally related to the cobalamin-binding domain of methionine synthase. Its corrinoid cofactor at the top of the Rossmann domain reaches deeply into the funnel of MtaB, defining a region between zinc(II) and the corrinoid cobalt that must be the binding site for methanol. The active site geometry supports a S(N)2 reaction mechanism, in which the C O bond in methanol is activated by the strong electrophile zinc(II) and cleaved because of an attack of the supernucleophile cob(I)amide. The environment of zinc(II) is characterized by an acidic cluster that increases the charge density on the zinc(II), polarizes methanol, and disfavors deprotonation of the methanol hydroxyl group. Implications of the MtaBC structure for the second step of the reaction, in which the methyl group is transferred to coenzyme M, are discussed.
- Max Planck Institute for Terrestrial Microbiology, Karl-von-Frisch-Strasse, D-35043 Marburg, Germany.
Organizational Affiliation: