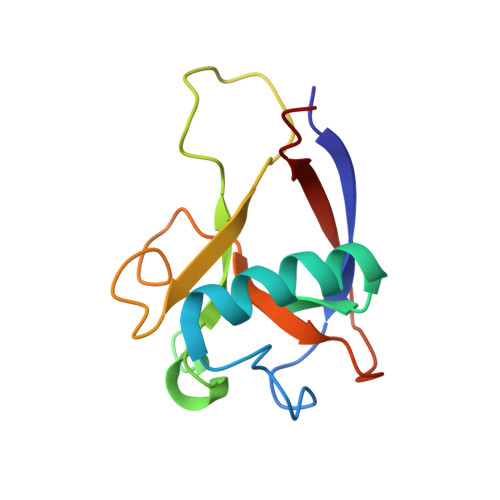
Novel beta-barrel fold in the nuclear magnetic resonance structure of the replicase nonstructural protein 1 from the severe acute respiratory syndrome coronavirus.
Almeida, M.S., Johnson, M.A., Herrmann, T., Geralt, M., Wuthrich, K.(2007) J Virol 81: 3151-3161
- PubMed: 17202208
- DOI: https://doi.org/10.1128/JVI.01939-06
- Primary Citation of Related Structures:
2GDT, 2HSX - PubMed Abstract:
The nonstructural protein 1 (nsp1) of the severe acute respiratory syndrome coronavirus has 179 residues and is the N-terminal cleavage product of the viral replicase polyprotein that mediates RNA replication and processing. The specific function of nsp1 is not known. Here we report the nuclear magnetic resonance structure of the nsp1 segment from residue 13 to 128, which represents a novel alpha/beta-fold formed by a mixed parallel/antiparallel six-stranded beta-barrel, an alpha-helix covering one opening of the barrel, and a 3(10)-helix alongside the barrel. We further characterized the full-length 179-residue protein and show that the polypeptide segments of residues 1 to 12 and 129 to 179 are flexibly disordered. The structure is analyzed in a search for possible correlations with the recently reported activity of nsp1 in the degradation of mRNA.
- Department of Molecular Biology, MB-44, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037.
Organizational Affiliation: