Comparison of the Structures of HIV-2 Protease Complexes in Three Crystal Space Groups with an HIV-1 Protease Complex Structure
Mulichak, A.M., Watenpaugh, K.D.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| HIV-2 PROTEASE | 99 | Human immunodeficiency virus 2 | Mutation(s): 0 | 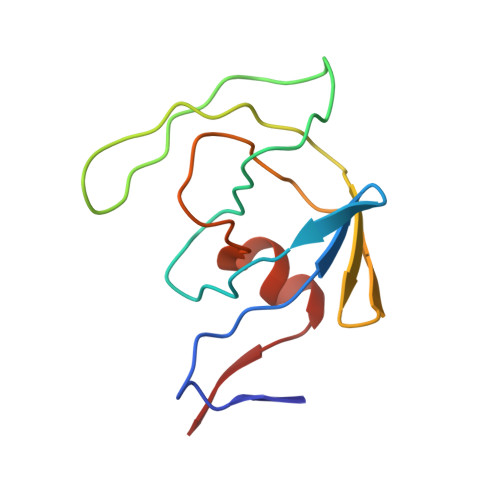 | |
UniProt | |||||
Find proteins for P04584 (Human immunodeficiency virus type 2 subtype A (isolate ROD)) Explore P04584 Go to UniProtKB: P04584 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P04584 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| UNIDENTIFIED PEPTIDE FRAGMENT | C [auth S] | 9 | N/A | Mutation(s): 0 |  |
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 58.23 | α = 90 |
| b = 43.82 | β = 106.16 |
| c = 39.18 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PROLSQ | refinement |