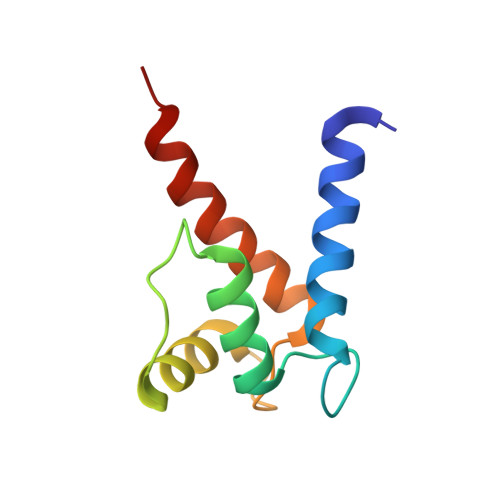
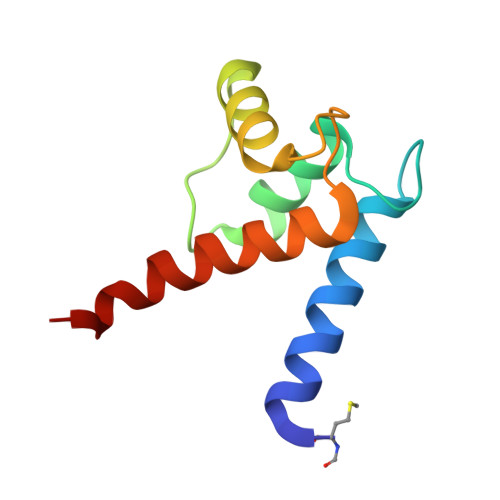
Structural and functional insights into RAGE activation by multimeric S100B.
Ostendorp, T., Leclerc, E., Galichet, A., Koch, M., Demling, N., Weigle, B., Heizmann, C.W., Kroneck, P.M., Fritz, G.(2007) EMBO J 26: 3868-3878
- PubMed: 17660747
- DOI: https://doi.org/10.1038/sj.emboj.7601805
- Primary Citation of Related Structures:
2H61 - PubMed Abstract:
Nervous system development and plasticity require regulation of cell proliferation, survival, neurite outgrowth and synapse formation by specific extracellular factors. The EF-hand protein S100B is highly expressed in human brain. In the extracellular space, it promotes neurite extension and neuron survival via the receptor RAGE (receptor for advanced glycation end products). The X-ray structure of human Ca(2+)-loaded S100B was determined at 1.9 A resolution. The structure revealed an octameric architecture of four homodimeric units arranged as two tetramers in a tight array. The presence of multimeric forms in human brain extracts was confirmed by size-exclusion experiments. Recombinant tetrameric, hexameric and octameric S100B were purified from Escherichia coli and characterised. Binding studies show that tetrameric S100B binds RAGE with higher affinity than dimeric S100B. Analytical ultracentrifugation studies imply that S100B tetramer binds two RAGE molecules via the V-domain. In line with these experiments, S100B tetramer caused stronger activation of cell growth than S100B dimer and promoted cell survival. The structural and the binding data suggest that tetrameric S100B triggers RAGE activation by receptor dimerisation.
- Fachbereich Biologie, Mathematisch-Naturwissenschaftliche Sektion, Universität Konstanz, Konstanz, Germany.
Organizational Affiliation: