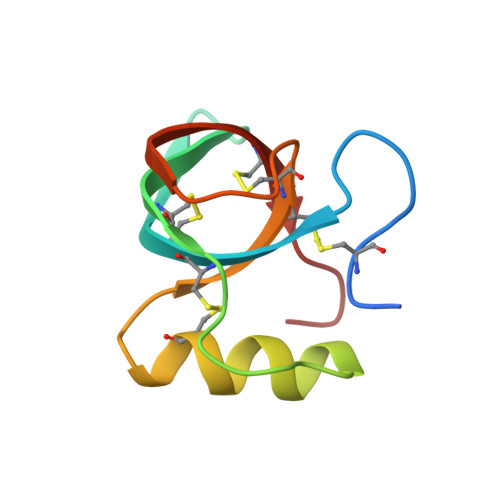
Two crystal structures of Trichoderma reesei hydrophobin HFBI--The structure of a protein amphiphile with and without detergent interaction.
Hakanpaa, J.M., Szilvay, G.R., Kaljunen, H., Maksimainen, M., Linder, M., Rouvinen, J.(2006) Protein Sci 15: 2129-2140
- PubMed: 16882996
- DOI: https://doi.org/10.1110/ps.062326706
- Primary Citation of Related Structures:
2FZ6, 2GVM - PubMed Abstract:
Hydrophobins are small fungal proteins that are highly surface active and possess a unique ability to form amphiphilic membranes through spontaneous self-assembly. The first crystal structure of a hydrophobin, Trichoderma reesei HFBII, revealed the structural basis for the function of this amphiphilic protein--a patch consisting of hydrophobic side chains on the protein surface. Here, the crystal structures of a native and a variant T. reesei hydrophobin HFBI are presented, revealing the same overall structure and functional hydrophobic patch as in the HFBII structure. However, some structural flexibility was found in the native HFBI structure: The asymmetric unit contained four molecules, and, in two of these, an area of seven residues was displaced as compared to the two other HFBI molecules and the previously determined HFBII structure. This structural change is most probably induced by multimer formation. Both the native and the N-Cys-variant of HFBI were crystallized in the presence of detergents, but an association between the protein and a detergent was only detected in the variant structure. There, the molecules were arranged into an extraordinary detergent-associated octamer and the solvent content of the crystals was 75%. This study highlights the conservation of the fold of class II hydrophobins in spite of the low sequence identity and supports our previous suggestion that concealment of the hydrophobic surface areas of the protein is the driving force in the formation of multimers and monolayers in the self-assembly process.
- Department of Chemistry, University of Joensuu, Finland.
Organizational Affiliation: