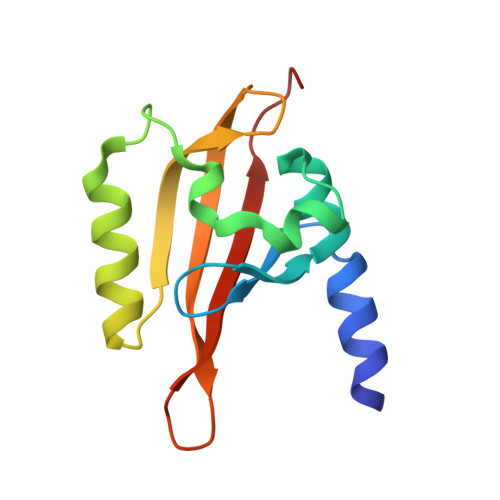
Structure of the redox sensor domain of Azotobacter vinelandii NifL at atomic resolution: signaling, dimerization, and mechanism.
Key, J., Hefti, M., Purcell, E.B., Moffat, K.(2007) Biochemistry 46: 3614-3623
- PubMed: 17319691
- DOI: https://doi.org/10.1021/bi0620407
- Primary Citation of Related Structures:
2GJ3 - PubMed Abstract:
NifL is a multidomain sensor protein responsible for the transcriptional regulation of genes involved in response to changes in cellular redox state and ADP concentration. Cellular redox is monitored by the N-terminal PAS domain of NifL which contains an FAD cofactor. Flavin-based PAS domains of this type have also been referred to as LOV domains. To explore the mechanism of signal recognition and transduction in NifL, we determined the crystal structure of the FAD-bound PAS domain of NifL from Azotobacter vinelandii to 1.04 A resolution. The structure reveals a novel cavity within the PAS domain which contains two water molecules directly coordinated to the FAD. This cavity is connected to solvent by multiple access channels which may facilitate the oxidation of the FAD by molecular oxygen and the release of hydrogen peroxide. The structure contains a dimer of the NifL PAS domain that is structurally very similar to those described in other crystal structures of PAS domains and identifies a conserved dimerization motif. An N-terminal amphipathic helix constitutes part of the dimerization interface, and similar N-terminal helices are identified in other PAS domain proteins. The structure suggests a model for redox-mediated signaling in which a conformational change is initiated by redox-dependent changes in protonation at the N5 atom of FAD that lead to reorganization of hydrogen bonds within the flavin binding pocket. A structural signal is subsequently transmitted to the beta-sheet interface between the monomers of the PAS domain.
- Department of Biochemistry and Molecular Biology, University of Chicago, Chicago, Illinois 60637, USA.
Organizational Affiliation: