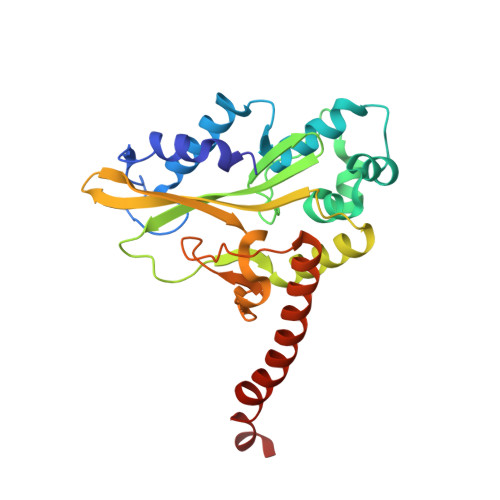
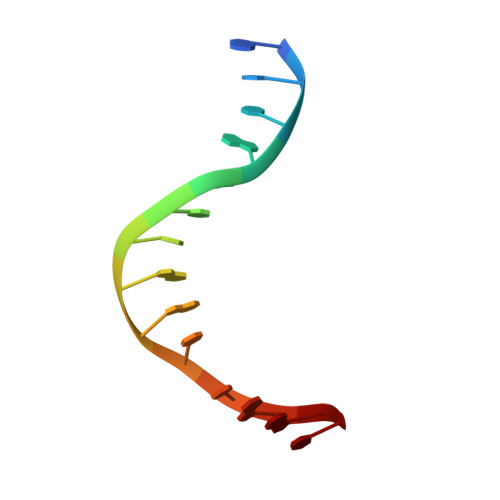
Alteration of Sequence Specificity of the Type II Restriction Endonuclease HincII through an Indirect Readout Mechanism.
Joshi, H.K., Etzkorn, C., Chatwell, L., Bitinaite, J., Horton, N.C.(2006) J Biological Chem 281: 23852-23869
- PubMed: 16675462
- DOI: https://doi.org/10.1074/jbc.M512339200
- Primary Citation of Related Structures:
2GIE, 2GIG, 2GIH, 2GII, 2GIJ - PubMed Abstract:
The functional and structural consequences of a mutation of the DNA intercalating residue of HincII, Q138F, are presented. Modeling has suggested that the DNA intercalation by Gln-138 results in DNA distortions potentially used by HincII in indirect readout of its cognate DNA, GTYRAC (Y = C or T, R = A or G) (Horton, N. C., Dorner, L. F., and Perona, J. J. (2002) Nat. Struct. Biol. 9, 42-47). Kinetic data presented here indicate that the mutation of glutamine 138 to phenylalanine (Q138F) results in a change in sequence specificity at the center two base pairs of the cognate recognition site. We show that the preference of HincII for cutting, but not binding, the three cognate sites differing in the center two base pairs has been altered by the mutation Q138F. Five new crystal structures are presented including Q138F HincII bound to GTTAAC and GTCGAC both with and without Ca2+ as well as the structure of wild type HincII bound to GTTAAC. The Q138F HincII/DNA structures show conformational changes in the protein, bound DNA, and at the protein-DNA interface, consistent with the formation of adaptive complexes. Analysis of these structures and the effect of Ca2+ binding on the protein-DNA interface illuminates the origin of the altered specificity by the mutation Q138F in the HincII enzyme.
- Department of Biochemistry and Molecular Biophysics, University of Arizona, Tucson, Arizona 85721, USA.
Organizational Affiliation: