Recombinant Thermus aquaticus RNA Polymerase for Structural Studies.
Kuznedelov, K., Lamour, V., Patikoglou, G., Chlenov, M., Darst, S.A., Severinov, K.(2006) J Mol Biology 359: 110-121
- PubMed: 16618493
- DOI: https://doi.org/10.1016/j.jmb.2006.03.009
- Primary Citation of Related Structures:
2GHO - PubMed Abstract:
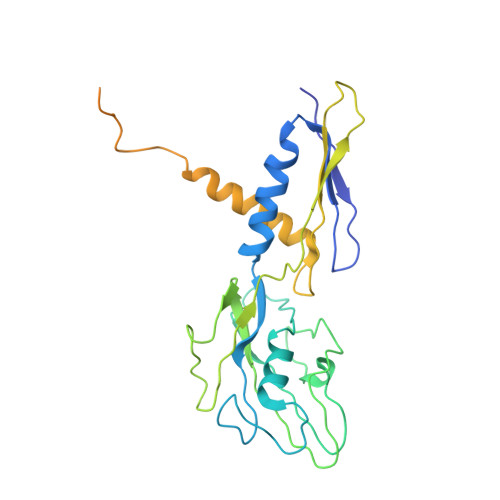
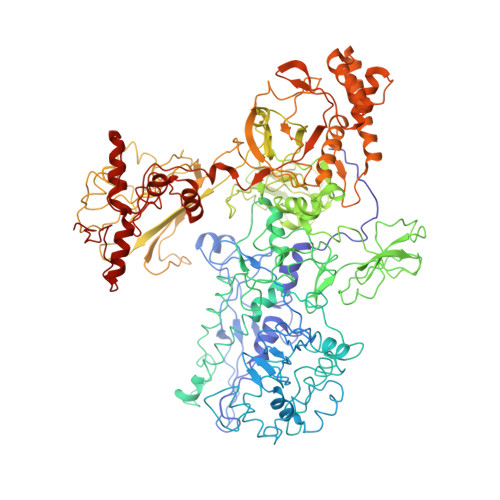
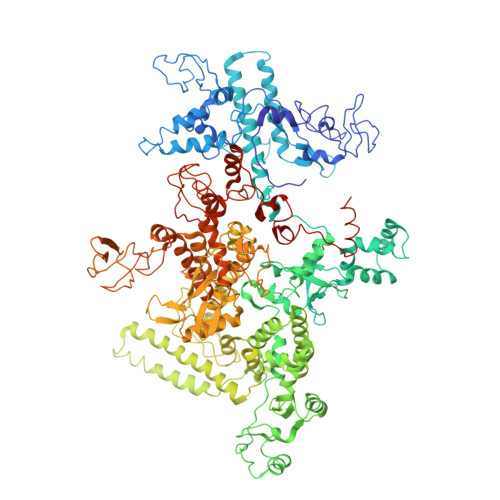
Advances in the structural biology of bacterial transcription have come from studies of RNA polymerases (RNAPs) from the thermophilic eubacteria Thermus aquaticus (Taq) and Thermus thermophilus (Tth). These structural studies have been limited by the fact that only endogenous Taq or Tth RNAP, laboriously purified from large quantities of Taq or Tth cell paste and offering few options for genetic modification, is suitable for structural studies. Recombinant systems for the preparation of Taq RNAP by co-overexpression and assembly in the heterologous host, Escherichia coli, have been described, but these did not yield enzyme suitable for crystallographic studies. Here we describe recombinant systems for the preparation of Taq RNAP harboring full or partial deletions of the Taq beta' non-conserved domain (NCD), yielding enzyme suitable for crystallographic studies. This opens the way for structural studies of genetically manipulated enzymes, allowing the preparation of more crystallizable enzymes and facilitating detailed structure/function analysis. Characterization of the Taqbeta'NCD deletion mutants generated in this study showed that the beta'NCD is important for the efficient binding of the sigma subunit, confirming previous hypotheses. Finally, preliminary structural analysis (at 4.1Angstroms resolution) of one of the recombinant mutants revealed a previously unobserved conformation of the beta-flap, further defining the range of conformations accessible to this flexible structural element.
- Department of Molecular Biology and Biochemistry, Rutgers University, Piscataway, NJ 08854, USA.
Organizational Affiliation: