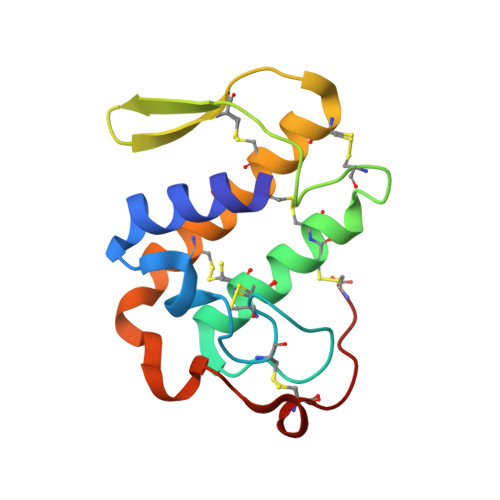
Crystal structure of a complex of phospholipase A2 with a designed peptide inhibitor Dehydro-Ile-Ala-Arg-Ser at 0.98 A resolution
Prem Kumar, R., Singh, N., Somvanshi, R.K., Ethayathulla, A.S., Dey, S., Sharma, S., Kaur, P., Perbandt, M., Betzel, C., Singh, T.P.To be published.