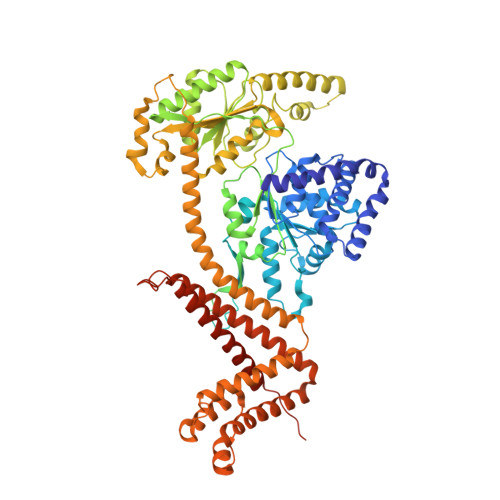
Structure of dimeric SecA, the Escherichia coli preprotein translocase motor.
Papanikolau, Y., Papadovasilaki, M., Ravelli, R.B., McCarthy, A.A., Cusack, S., Economou, A., Petratos, K.(2007) J Mol Biology 366: 1545-1557
- PubMed: 17229438
- DOI: https://doi.org/10.1016/j.jmb.2006.12.049
- Primary Citation of Related Structures:
2FSF, 2FSG, 2FSH, 2FSI - PubMed Abstract:
SecA is the preprotein translocase ATPase subunit and a superfamily 2 (SF2) RNA helicase. Here we present the 2 A crystal structures of the Escherichia coli SecA homodimer in the apo form and in complex with ATP, ADP and adenosine 5'-[beta,gamma-imido]triphosphate (AMP-PNP). Each monomer contains the SF2 ATPase core (DEAD motor) built of two domains (nucleotide binding domain, NBD and intramolecular regulator of ATPase 2, IRA2), the preprotein binding domain (PBD), which is inserted in NBD and a carboxy-terminal domain (C-domain) linked to IRA2. The structures of the nucleotide complexes of SecA identify an interfacial nucleotide-binding cleft located between the two DEAD motor domains and residues critical for ATP catalysis. The dimer comprises two virtually identical protomers associating in an antiparallel fashion. Dimerization is mediated solely through extensive contacts of the DEAD motor domains leaving the C-domain facing outwards from the dimerization core. This dimerization mode explains the effect of functionally important mutations and is completely different from the dimerization models proposed for other SecA structures. The repercussion of these findings on translocase assembly and catalysis is discussed.
- Institute of Molecular Biology and Biotechnology, Foundation for Research and Technology, PO Box 1385, 71110 Heraklion, Greece.
Organizational Affiliation: