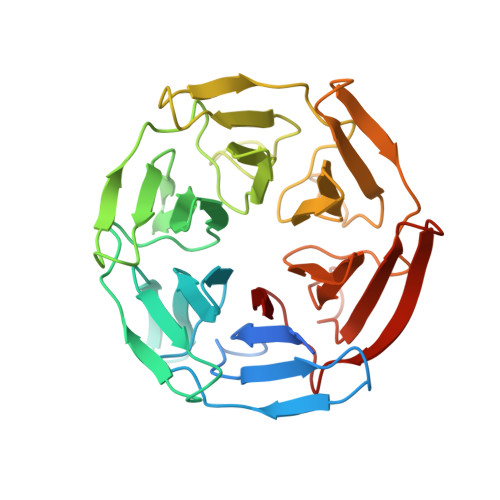
Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling.
Lo, S.C., Li, X., Henzl, M.T., Beamer, L.J., Hannink, M.(2006) EMBO J 25: 3605-3617
- PubMed: 16888629
- DOI: https://doi.org/10.1038/sj.emboj.7601243
- Primary Citation of Related Structures:
2FLU - PubMed Abstract:
Keap1 is a BTB-Kelch substrate adaptor protein that regulates steady-state levels of Nrf2, a bZIP transcription factor, in response to oxidative stress. We have determined the structure of the Kelch domain of Keap1 bound to a 16-mer peptide from Nrf2 containing a highly conserved DxETGE motif. The Nrf2 peptide contains two short antiparallel beta-strands connected by two overlapping type I beta-turns stabilized by the aspartate and threonine residues. The beta-turn region fits into a binding pocket on the top face of the Kelch domain and the glutamate residues form multiple hydrogen bonds with highly conserved residues in Keap1. Mutagenesis experiments confirmed the role of individual amino acids for binding of Nrf2 to Keap1 and for Keap1-mediated repression of Nrf2-dependent gene expression. Our results provide a detailed picture of how a BTB-Kelch substrate adaptor protein binds to its cognate substrate and will enable the rational design of novel chemopreventive agents.
- Department of Biochemistry, University of Missouri-Columbia, Columbia, MO 65211, USA.
Organizational Affiliation: