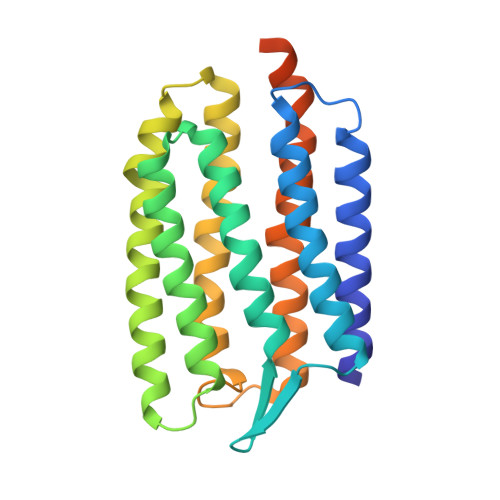
Development of the signal in sensory rhodopsin and its transfer to the cognate transducer.
Moukhametzianov, R., Klare, J.P., Efremov, R., Baeken, C., Goppner, A., Labahn, J., Engelhard, M., Buldt, G., Gordeliy, V.I.(2006) Nature 440: 115-119
- PubMed: 16452929
- DOI: https://doi.org/10.1038/nature04520
- Primary Citation of Related Structures:
2F93, 2F95 - PubMed Abstract:
The microbial phototaxis receptor sensory rhodopsin II (NpSRII, also named phoborhodopsin) mediates the photophobic response of the haloarchaeon Natronomonas pharaonis by modulating the swimming behaviour of the bacterium. After excitation by blue-green light NpSRII triggers, by means of a tightly bound transducer protein (NpHtrII), a signal transduction chain homologous with the two-component system of eubacterial chemotaxis. Two molecules of NpSRII and two molecules of NpHtrII form a 2:2 complex in membranes as shown by electron paramagnetic resonance and X-ray structure analysis. Here we present X-ray structures of the photocycle intermediates K and late M (M2) explaining the evolution of the signal in the receptor after retinal isomerization and the transfer of the signal to the transducer in the complex. The formation of late M has been correlated with the formation of the signalling state. The observed structural rearrangements allow us to propose the following mechanism for the light-induced activation of the signalling complex. On excitation by light, retinal isomerization leads in the K state to a rearrangement of a water cluster that partly disconnects two helices of the receptor. In the transition to late M the changes in the hydrogen bond network proceed further. Thus, in late M state an altered tertiary structure establishes the signalling state of the receptor. The transducer responds to the activation of the receptor by a clockwise rotation of about 15 degrees of helix TM2 and a displacement of this helix by 0.9 A at the cytoplasmic surface.
- Research Centre Jülich, Institute of Structural Biology (IBI-2), 52425 Jülich, Germany.
Organizational Affiliation: