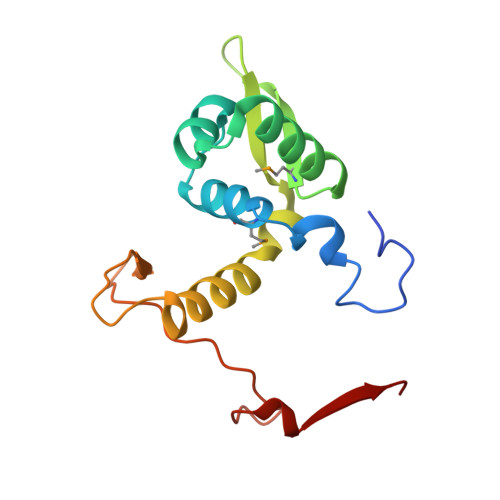
The X-ray crystal structure of PA1607 from Pseudomonas aureginosa at 1.9 A resolution--a putative transcription factor.
Sieminska, E.A., Xu, X., Savchenko, A., Sanders, D.A.(2007) Protein Sci 16: 543-549
- PubMed: 17322537
- DOI: https://doi.org/10.1110/ps.062668207
- Primary Citation of Related Structures:
2F2E - PubMed Abstract:
The structure of the PA1607 protein from Pseudomonas aureginosa was determined at 1.85 A resolution using the Se-Met multiwavelength anomalous diffraction (MAD) technique. PA1607 forms a dimer and adopts a winged-helix motif similar to the MarR family of transcription regulators, though it has an unusual dimerization profile. The DNA-binding regions and a putative metal-binding site are not conserved in PA1607.
- Department of Chemistry, University of Saskatchewan, Saskatoon, Saskatchewan S7N 5C9, Canada.
Organizational Affiliation: