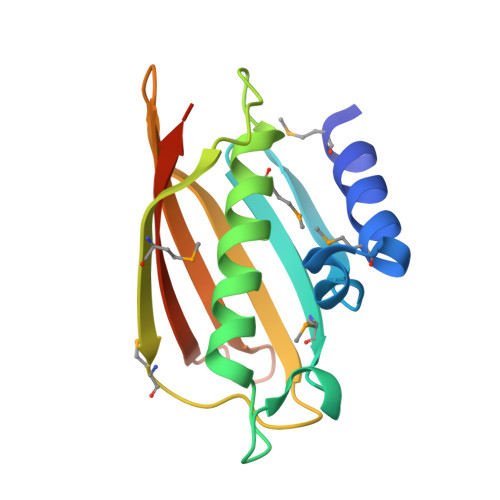
Crystal structure of human thioesterase superfamily member 2
Cheng, Z., Song, F., Shan, X., Wei, Z., Wang, Y., Dunaway-Mariano, D., Gong, W.(2006) Biochem Biophys Res Commun 349: 172-177
- PubMed: 16934754
- DOI: https://doi.org/10.1016/j.bbrc.2006.08.025
- Primary Citation of Related Structures:
2F0X - PubMed Abstract:
Hotdog-fold has been identified in more than 1000 proteins, yet many of which in eukaryotes are less studied. No structural or functional studies of human thioesterase superfamily member 2 (hTHEM2) have been reported before. Since hTHEM2 exhibits about 20% sequence identity to Escherichia coli PaaI protein, it was proposed to be a thioesterase with a hotdog-fold. Here, we report the crystallographic structure of recombinant hTHEM2, determined by the single-wavelength anomalous dispersion method at 2.3A resolution. This structure demonstrates that hTHEM2 indeed contains a hotdog-fold and forms a back-to-back tetramer as other hotdog proteins. Based on structural and sequence conservation, the thioesterase active site in hTHEM2 is predicted. The structure and substrate specificity are most similar to those of the bacterial phenylacetyl-CoA hydrolase. Asp65, located on the central alpha-helix of subunit B, was shown by site-directed mutagenesis to be essential to catalysis.
- School of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230026, PR China.
Organizational Affiliation: