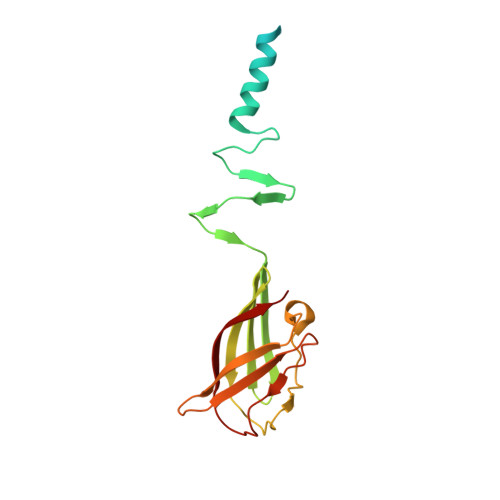
Modular structure of the receptor binding proteins of Lactococcus lactis phages. The RBP structure of the temperate phage TP901-1.
Spinelli, S., Campanacci, V., Blangy, S., Moineau, S., Tegoni, M., Cambillau, C.(2006) J Biological Chem 281: 14256-14262
- PubMed: 16549427
- DOI: https://doi.org/10.1074/jbc.M600666200
- Primary Citation of Related Structures:
2F0C - PubMed Abstract:
Lactococcus lactis is a gram-positive bacterium widely used by the dairy industry. Several industrial L. lactis strains are sensitive to various distinct bacteriophages. Most of them belong to the Siphoviridae family and comprise several species, among which the 936 and P335 are prominent. Members of these two phage species recognize their hosts through the interaction of their receptor-binding protein (RBP) with external cell wall saccharidices of the host, the "receptors." We report here the 1.65 A resolution crystal structure of the RBP from phage TP901-1, a member of the P335 species. This RBP of 163 amino acids is a homotrimer comprising three domains: a helical N terminus, an interlaced beta-prism, and a beta-barrel, the head domain (residues 64-163), which binds a glycerol molecule. Fluorescence quenching experiments indicated that the RBP exhibits high affinity for glycerol, muramyl-dipeptide, and other saccharides in solution. The structural comparison of this RBP with that of lactococcal phage p2 RBP, a member of the 936 species (Spinelli, S., Desmyter, A., Verrips, C. T., de Haard, J. W., Moineau, S., and Cambillau, C. (2006) Nat. Struct. Mol. Biol. 13, 85-89) suggests a large extent of modularity in RBPs of lactococcal phages.
- Architecture et Fonction des Macromolécules Biologiques, UMR 6098 CNRS and Université d'Aix-Marseille I, Campus de Luminy, Case 932, 13288 Marseille Cedex 09, France.
Organizational Affiliation: