Regulation through the RNA Polymerase Secondary Channel: STRUCTURAL AND FUNCTIONAL VARIABILITY OF THE COILED-COIL TRANSCRIPTION FACTORS.
Symersky, J., Perederina, A., Vassylyeva, M.N., Svetlov, V., Artsimovitch, I., Vassylyev, D.G.(2006) J Biological Chem 281: 1309-1312
- PubMed: 16298991
- DOI: https://doi.org/10.1074/jbc.C500405200
- Primary Citation of Related Structures:
2EUL - PubMed Abstract:
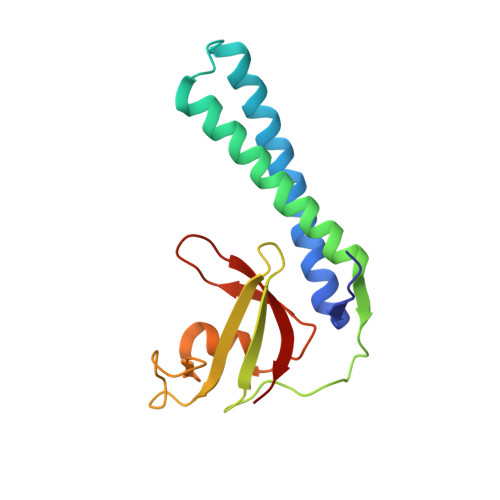
Gre factors enhance the intrinsic endonucleolytic activity of RNA polymerase to rescue arrested transcription complexes and are thought to confer the high fidelity and processivity of RNA synthesis. The Gre factors insert the extended alpha-helical coiled-coil domains into the RNA polymerase secondary channel to position two invariant acidic residues at the coiled-coil tip near the active site to stabilize the catalytic metal ion. Gfh1, a GreA homolog from Thermus thermophilus, inhibits rather than activates RNA cleavage. Here we report the structure of the T. thermophilus Gfh1 at 2.4 A resolution revealing a two-domain architecture closely resembling that of GreA. However, the interdomain orientation is strikingly distinct (approximately 162 degrees rotation) between the two proteins. In contrast to GreA, which has two acidic residues on a well fixed self-stabilized alpha-turn, the tip of the Gfh1 coiled-coil is flexible and contains four acidic residues. This difference is likely the key to the Gre functional diversity, while Gfh1 inhibits exo- and endonucleolytic cleavage, RNA synthesis, and pyrophosphorolysis, GreA enhances only the endonucleolytic cleavage. We propose that Gfh1 acidic residues stabilize the RNA polymerase active center in a catalytically inactive configuration through Mg2+-mediated interactions. The excess of the acidic residues and inherent flexibility of the coiled-coil tip might allow Gfh1 to adjust its activity to structurally distinct substrates, thereby inhibiting diverse catalytic reactions of RNA polymerase.
- Department of Biochemistry and Molecular Genetics, University of Alabama at Birmingham, School of Medicine, Birmingham, Alabama 35294, USA.
Organizational Affiliation: