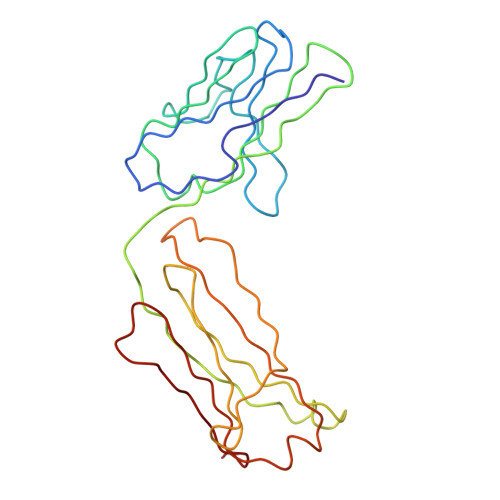
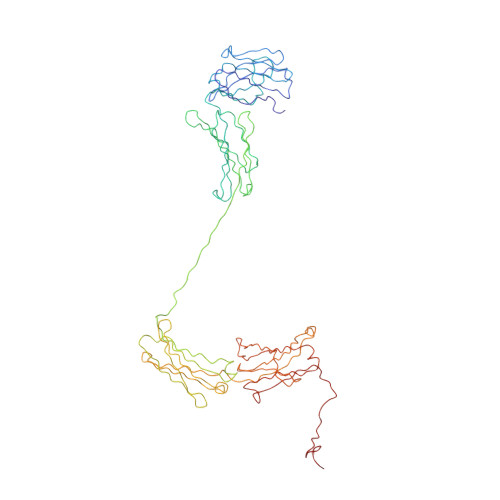
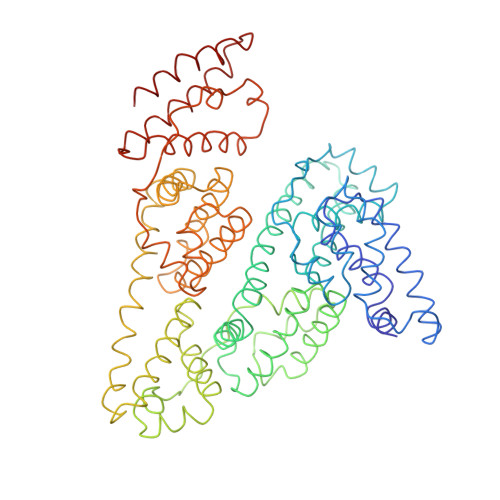
Purification, Properties and Extended Solution Structure of the Complex Formed between Human Immunoglobulin A1 and Human Serum Albumin by Scattering and Ultracentrifugation.
Almogren, A., Furtado, P.B., Sun, Z., Perkins, S.J., Kerr, M.A.(2006) J Mol Biology 356: 413-431
- PubMed: 16376934
- DOI: https://doi.org/10.1016/j.jmb.2005.11.060
- Primary Citation of Related Structures:
2ESG - PubMed Abstract:
Immunoglobulin A (IgA) is unique amongst antibodies in being able to form polymeric structures that may possess important functions in the pathology of specific diseases. IgA also forms complexes with other plasma proteins, the IgA1-human serum albumin (HSA) complex (IgA1-HSA) being typical. We have purified this complex using a novel two-step purification based on thiophilic chromatography and gel filtration, and characterised this. HSA is linked covalently to the tailpiece of IgA1 by a disulphide bond between Cys471 in IgA1 and Cys34 in HSA. IgA1-HSA binds to IgA receptors on neutrophils and monocytes, and elicits a respiratory burst that is comparable in magnitude to that of monomeric IgA1. The solution arrangement of IgA1-HSA was identified by X-ray scattering and ultracentrifugation. The radius of gyration R(G) of 7.5(+/-0.3) nm showed that IgA1-HSA is more extended in solution than IgA1 (R(G) of 6.1-6.2 nm). Its distance distribution function P(r) showed two peaks that indicated a well-separated solution structure similar to that for IgA1, and a maximum dimension of 25 nm, which is greater than that of 21 nm for IgA1. Sedimentation equilibrium showed that the IgA1:HSA stoichiometry is 1:1. Sedimentation velocity resulted in a sedimentation coefficient of 6.4S and a frictional ratio of 1.87, which is greater than that of 1.56 for IgA1. The constrained modelling of the IgA1-HSA structure using known structures for IgA1 and HSA generated 2432 conformationally randomised models of which 52 gave good scattering fits. The HSA structure was located at the base of the Fc fragment in IgA1 in an extended arrangement. Such a structure accounts for the functional activity of IgA1-HSA, and supports our previous modelling analysis of the IgA1 solution structure. The IgA1-HSA complex may suggest the potential for creating a new class of targeted therapeutic reagents based on the coupling of IgA1 to carrier proteins.
- Department of Pathology, College of Medicine and King Khalid University Hospital, King Saud University, Riyadh, Saudi Arabia.
Organizational Affiliation: