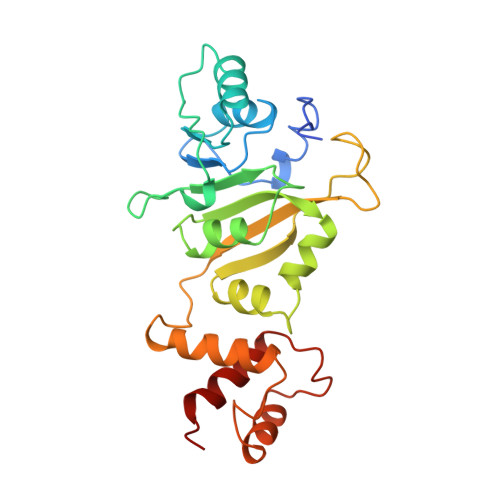
Crystal structure of ErmC', an rRNA methyltransferase which mediates antibiotic resistance in bacteria.
Bussiere, D.E., Muchmore, S.W., Dealwis, C.G., Schluckebier, G., Nienaber, V.L., Edalji, R.P., Walter, K.A., Ladror, U.S., Holzman, T.F., Abad-Zapatero, C.(1998) Biochemistry 37: 7103-7112
- PubMed: 9585521
- DOI: https://doi.org/10.1021/bi973113c
- Primary Citation of Related Structures:
2ERC - PubMed Abstract:
The prevalent mechanism of bacterial resistance to erythromycin and other antibiotics of the macrolide-lincosamide-streptogramin B group (MLS) is methylation of the 23S rRNA component of the 50S subunit in bacterial ribosomes. This sequence-specific methylation is catalyzed by the Erm group of methyltransferases (MTases). They are found in several strains of pathogenic bacteria, and ErmC is the most studied member of this class. The crystal structure of ErmC' (a naturally occurring variant of ErmC) from Bacillus subtilis has been determined at 3.0 A resolution by multiple anomalous diffraction phasing methods. The structure consists of a conserved alpha/beta amino-terminal domain which binds the cofactor S-adenosyl-l-methionine (SAM), followed by a smaller, alpha-helical RNA-recognition domain. The beta-sheet structure of the SAM-binding domain is well-conserved between the DNA, RNA, and small-molecule MTases. However, the C-terminal nucleic acid binding domain differs from the DNA-binding domains of other MTases and is unlike any previously reported RNA-recognition fold. A large, positively charged, concave surface is found at the interface of the N- and C-terminal domains and is proposed to form part of the protein-RNA interaction surface. ErmC' exhibits the conserved structural motifs previously found in the SAM-binding domain of other methyltransferases. A model of SAM bound to ErmC' is presented which is consistent with the motif conservation among MTases.
- Laboratory of Protein Crystallography, Department of Scientific Information, Analysis and Management, Abbott Laboratories, Abbott Park, Illinois 60064-3500, USA.
Organizational Affiliation: