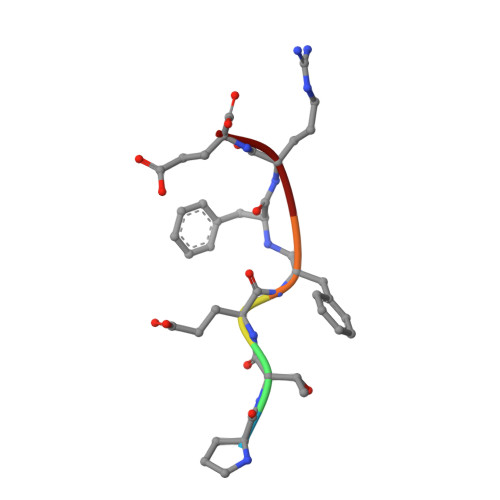
The structure of a synthetic pepsin inhibitor complexed with endothiapepsin
Cooper, J., Foundling, S., Hemmings, A., Blundell, T., Jones, D.M., Hallett, A., Szelke, M.(1987) Eur J Biochem 169: 215-221
- PubMed: 3119339
- DOI: https://doi.org/10.1111/j.1432-1033.1987.tb13600.x
- Primary Citation of Related Structures:
2ER6 - PubMed Abstract:
The conformation of a synthetic polypeptide inhibitor, bound to the active site of the fungal aspartic proteinase endothiapepsin (EC 3.4.23.6), has been determined by X-ray diffraction at 0.20-nm resolution and refined to an agreement factor of 0.20. The inhibitor: Pro Thr Glu Phe-R-Phe Arg Glu (R = -CH2NH-) is based on a chromogenic substrate of pepsin (EC 3.4.23.1). It has, in place of the scissile bond, a reduced peptide group which is resistant to hydrolysis and mimics the tetrahedral transition state. The inhibitor binds in an extended conformation with the reduced bond close to the essential aspartate side-chains of the enzyme. The hydrogen bonds and hydrophobic interactions between the enzyme and the inhibitor do not induce large conformational changes.
- Department of Crystallography, Birkbeck College, London, England.
Organizational Affiliation: