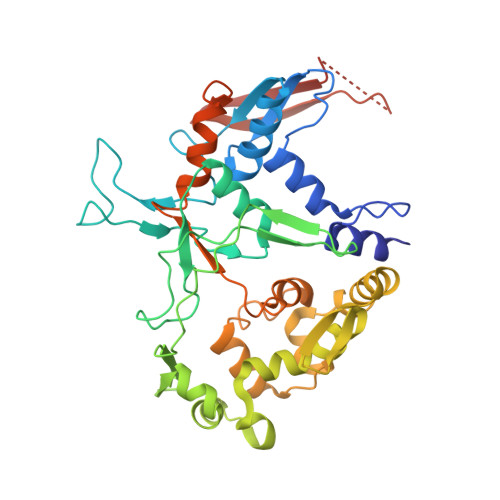
Structural characterization of l-glutamate oxidase from Streptomyces sp. X-119-6
Arima, J., Sasaki, C., Sakaguchi, C., Mizuno, H., Tamura, T., Kashima, A., Kusakabe, H., Sugio, S., Inagaki, K.(2009) FEBS J 276: 4318-4327
- PubMed: 19531050
- DOI: https://doi.org/10.1111/j.1742-4658.2009.07103.x
- Primary Citation of Related Structures:
2E1M - PubMed Abstract:
L-Glutamate oxidase (LGOX) from Streptomyces sp. X-119-6, which catalyzes the oxidative deamination of L-glutamate, has attracted increasing attention as a component of amperometric L-glutamate sensors used in the food industry and clinical biochemistry. The precursor of LGOX, which has a homodimeric structure, is less active than the mature enzyme with an alpha(2)beta(2)V(2) structure; enzymatic proteolysis of the precursor forms the stable mature enzyme. We solved the crystal structure of mature LGOX using molecular replacement with a structurally homologous model of L-amino acid oxidase (LAAO) from snake venom: LGOX has a deeply buried active site and two entrances from the surface of the protein into the active site. Comparison of the LGOX structure with that of LAAO revealed that LGOX has three regions that are absent from the LAAO structure, one of which is involved in the formation of the entrance. Furthermore, the arrangement of the residues composing the active site differs between LGOX and LAAO, and the active site of LGOX is narrower than that of LAAO. Results of the comparative analyses described herein raise the possibility that such a unique structure of LGOX is associated with its substrate specificity.
- Department of Biofunctional Chemistry, Graduate School of Natural Science and Technology, Okayama University, Japan.
Organizational Affiliation: