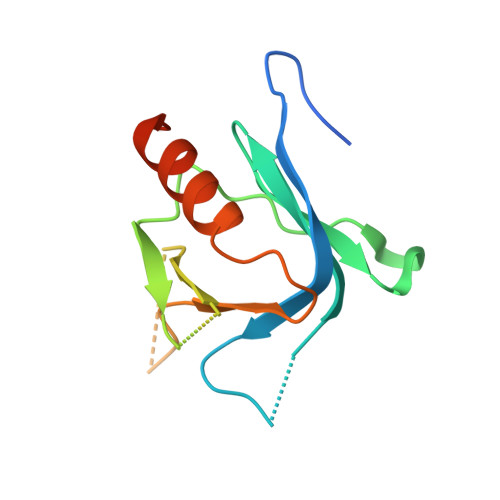
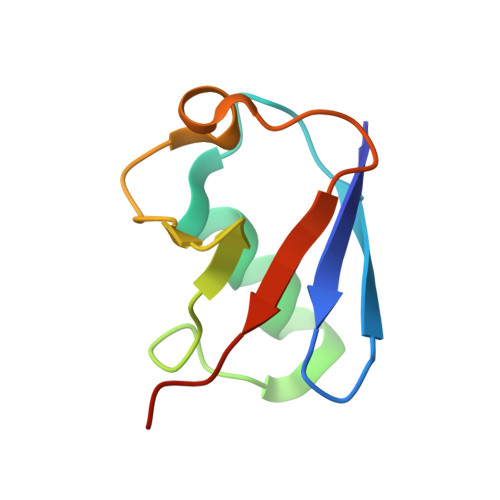
Structural basis of ubiquitin recognition by mammalian Eap45 GLUE domain
Hirano, S., Suzuki, N., Slagsvold, T., Kawasaki, M., Trambaiolo, D., Kato, R., Stenmark, H., Wakatsuki, S.(2006) Nat Struct Mol Biol 13: 1031-1032
- PubMed: 17057714
- DOI: https://doi.org/10.1038/nsmb1163
- Primary Citation of Related Structures:
2DX5 - PubMed Abstract:
ESCRT-II, a complex that sorts ubiquitinated membrane proteins to lysosomes, localizes to endosomes through interaction between the Vps36 subunit's GLUE domain and phosphatidylinositides (PIs). In yeast, a ubiquitin (Ub)-interacting NZF domain is inserted in Vps36 GLUE, whereas its mammalian counterpart, Eap45 GLUE, lacks the NZF domain. In the Eap45 GLUE-Ub complex structure, Ub binds far from the proposed PI-binding site of Eap45 GLUE, suggesting their independent binding.
- Structural Biology Research Center, Photon Factory, Institute of Materials Structure Science, High Energy Accelerator Research Organization (KEK), Tsukuba, Ibaraki 305-0801, Japan.
Organizational Affiliation: