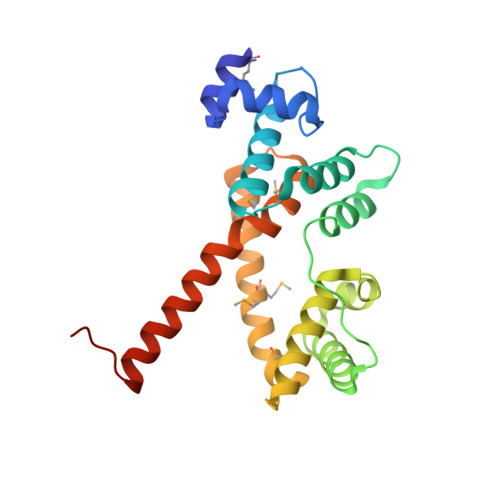
Structural and genomic DNA analysis of a putative transcription factor SCO5550 from Streptomyces coelicolor A3(2): regulating the expression of gene sco5551 as a transcriptional activator with a novel dimer shape
Hayashi, T., Tanaka, Y., Sakai, N., Watanabe, N., Tamura, T., Tanaka, I., Yao, M.(2013) Biochem Biophys Res Commun 435: 28-33
- PubMed: 23618855
- DOI: https://doi.org/10.1016/j.bbrc.2013.04.017
- Primary Citation of Related Structures:
2DG6 - PubMed Abstract:
SCO5550 from the model actinomycete Streptomyces coelicolor A3(2) was identified as a putative transcriptional regulator, and classified into the MerR family by sequence analysis. Recombined SCO5550 was successfully produced in Rhodococcus erythropolis, which can be used to stably express recombinant protein by optimizing the temperature over a wide range (4-35 °C). Crystal structure analysis showed that the dimerization domain (C-terminal domain) of SCO5550 has a novel fold and forms a new dimer shape, whereas the DNA-binding domain (N-terminal domain) is very similar to those of MerR family members. Such the new dimer form suggests that SCO5550 may define a new subfamily as a new member of the MerR family. Binding DNA sequence analysis of SCO5550 using the genomic systematic evolution of ligands by exponential enrichment (gSELEX) and electrophoretic mobility shift assay (EMSA) indicated that SCO5550 regulates the expression of the immediately upstream gene sco5551 encoding a putative protein, probably as a transcriptional activator.
- Department of Food and Fermentation Science, Faculty of Food and Nutrition, Beppu University, Beppu 874-8501, Japan.
Organizational Affiliation: