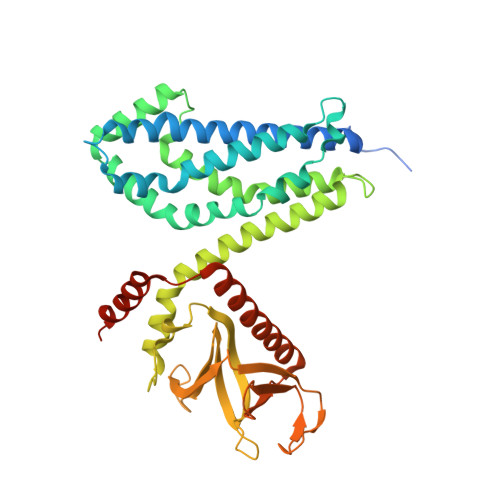
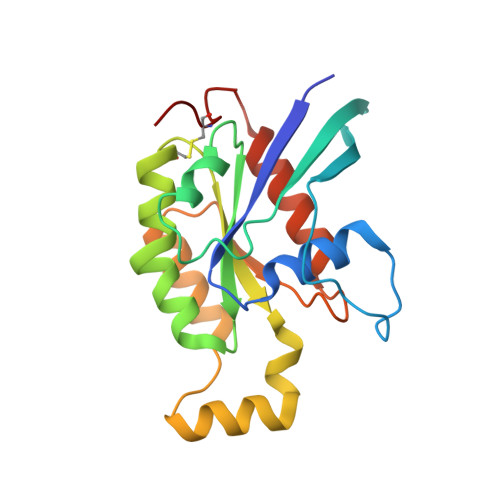
The Crystal Structure of Cdc42 in Complex with Collybistin II, a Gephyrin-interacting Guanine Nucleotide Exchange Factor.
Xiang, S., Kim, E.Y., Connelly, J.J., Nassar, N., Kirsch, J., Winking, J., Schwarz, G., Schindelin, H.(2006) J Mol Biology 359: 35-46
- PubMed: 16616186
- DOI: https://doi.org/10.1016/j.jmb.2006.03.019
- Primary Citation of Related Structures:
2DFK - PubMed Abstract:
The synaptic localization of ion channel receptors is essential for efficient synaptic transmission and the precise regulation of diverse neuronal functions. In the central nervous system, ion channel receptors reside in the postsynaptic membrane where they are juxtaposed to presynaptic terminals. For proper function, these ion channels have to be anchored to the cytoskeleton, and in the case of the inhibitory glycine and gamma-amino-butyric acid type A (GABA(A)) receptors this interaction is mediated by a gephyrin centered scaffold. Highlighting its central role in this receptor anchoring scaffold, gephyrin interacts with a number of proteins, including the neurospecific guanine nucleotide exchange factor collybistin. Collybistin belongs to the Dbl family of guanine nucleotide exchange factors, occurs in multiple splice variants, and is specific for Cdc42, a small GTPase belonging to the Rho family. The 2.3 Angstroms resolution crystal structure of the Cdc42-collybistin II complex reveals a novel conformation of the switch I region of Cdc42. It also provides the first direct observation of structural changes in the relative orientation of the Dbl-homology domain and the pleckstrin-homology domain in the same Dbl family protein. Biochemical data indicate that gephyrin negatively regulates collybistin activity.
- Department of Biochemistry, SUNY Stony Brook, NY 11794-5215, USA.
Organizational Affiliation: