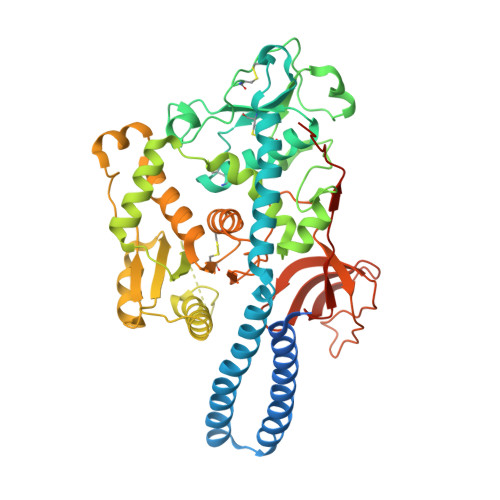
Crystal structure of mammalian {alpha}1,6-fucosyltransferase, FUT8
Ihara, H., Ikeda, Y., Toma, S., Wang, X., Suzuki, T., Gu, J., Miyoshi, E., Tsukihara, T., Honke, K., Matsumoto, A., Nakagawa, A., Taniguchi, N.(2007) Glycobiology 17: 455-466
- PubMed: 17172260
- DOI: https://doi.org/10.1093/glycob/cwl079
- Primary Citation of Related Structures:
2DE0 - PubMed Abstract:
Mammalian alpha1,6-fucosyltransferase (FUT8) catalyses the transfer of a fucose residue from a donor substrate, guanosine 5'-diphosphate-beta-L-fucose to the reducing terminal N-acetylglucosamine (GlcNAc) of the core structure of an asparagine-linked oligosaccharide. Alpha1,6-fucosylation, also referred to as core fucosylation, plays an essential role in various pathophysiological events. Our group reported that FUT8 null mice showed severe growth retardation and emphysema-like lung-destruction as a result of the dysfunction of epidermal growth factor and transforming growth factor-beta receptors. To elucidate the molecular basis of FUT8 with respect to pathophysiology, the crystal structure of human FUT8 was determined at 2.6 A resolution. The overall structure of FUT8 was found to consist of three domains: an N-terminal coiled-coil domain, a catalytic domain, and a C-terminal SH3 domain. The catalytic region appears to be similar to GT-B glycosyltransferases rather than GT-A. The C-terminal part of the catalytic domain of FUT8 includes a Rossmann fold with three regions that are conserved in alpha1,6-, alpha1,2-, and protein O-fucosyltransferases. The SH3 domain of FUT8 is similar to other SH3 domain-containing proteins, although the significance of this domain remains to be elucidated. The present findings of FUT8 suggest that the conserved residues in the three conserved regions participate in the Rossmann fold and act as the donor binding site, or in catalysis, thus playing key roles in the fucose-transferring reaction.
- Department of Disease Glycomics, Research Institute for Microbial Diseases, Osaka University, Taniguchi Research Group, 4th Floor, Center for Advanced Science & Innovation, Osaka University, 2-1, Yamadaoka Suita, Osaka 565-0871, Japan.
Organizational Affiliation: