Structural insights into the cryptic DNA-dependent ATPase activity of UvrB
Eryilmaz, J., Ceschini, S., Ryan, J., Geddes, S., Waters, T.R., Barrett, T.E.(2006) J Mol Biology 357: 62-72
- PubMed: 16426634
- DOI: https://doi.org/10.1016/j.jmb.2005.12.059
- Primary Citation of Related Structures:
2D7D - PubMed Abstract:
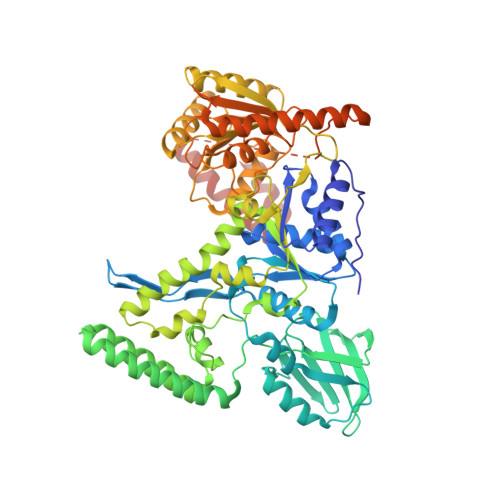
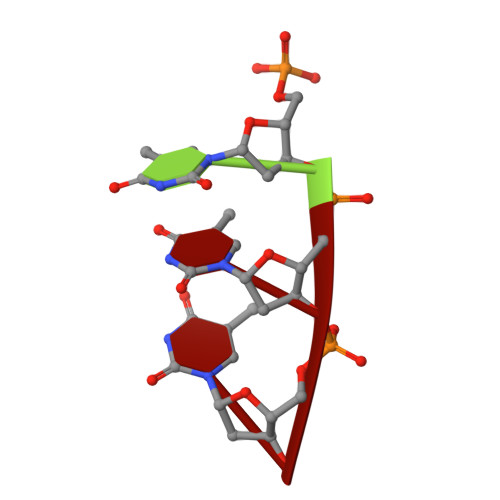
The UvrABC pathway is a ubiquitously occurring mechanism targeted towards the repair of bulky base damage. Key to this process is UvrB, a DNA-dependent limited helicase that acts as a lesion recognition element whilst part of a tracking complex involving UvrA, and as a DNA-binding platform required for the presentation of damage to UvrC for subsequent processing. We have been able to determine the structure of a ternary complex involving UvrB* (a C-terminal truncation of full-length UvrB), a polythymine trinucleotide and ADP. This structure has highlighted the roles of key conserved residues in DNA binding distinct from those of the beta-hairpin, where most of the attention in previous studies has been focussed. We are also the first to report the structural basis underlying conformational re-modelling of the beta-hairpin that is absolutely required for DNA binding and how this event results in an ATPase primed for catalysis. Our data provide the first insights at the molecular level into the transformation of UvrB into an active helicase.
- The School of Crystallography and the Institute for Structural Molecular Biology, Birkbeck College, Malet Street, London WC1E 7HX, UK.
Organizational Affiliation: