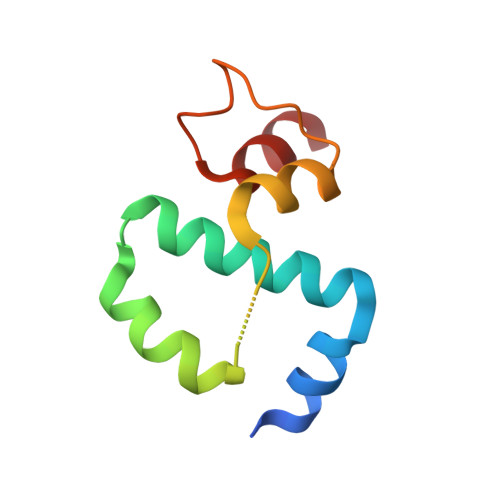
Structure of the N-terminal Domain of the FOP (FGFR1OP) Protein and Implications for its Dimerization and Centrosomal Localization.
Mikolajka, A., Yan, X., Popowicz, G.M., Smialowski, P., Nigg, E.A., Holak, T.A.(2006) J Mol Biology 359: 863-875
- PubMed: 16690081
- DOI: https://doi.org/10.1016/j.jmb.2006.03.070
- Primary Citation of Related Structures:
2D68 - PubMed Abstract:
The fibroblast growth factor receptor 1 (FGFR1) oncogene partner, FOP, is a centrosomal protein that is involved in the anchoring of microtubules (MTS) to subcellular structures. The protein was originally discovered as a fusion partner with FGFR1 in oncoproteins that give rise to stem cell myeloproliferative disorders. A subsequent proteomics screen identified FOP as a component of the centrosome. FOP contains a Lis-homology (LisH) motif found in more than 100 eukaryotic proteins. LisH motifs are believed to be involved in microtubule dynamics and organization, cell migration, and chromosome segregation; several of them are associated with genetic diseases. We report here a 1.6A resolution crystal structure of the N-terminal dimerization domain of FOP. The structure comprises an alpha-helical bundle composed of two antiparallel chains, each of them having five alpha-helices. The central part of the dimer contains the LisH domain. We further determined that the FOP LisH domain is part of a longer N-terminal segment that is required, albeit not sufficient, for dimerization and centrosomal localization of FOP.
- Max Planck Institute of Biochemistry, D-82152 Martinsried, Germany.
Organizational Affiliation: