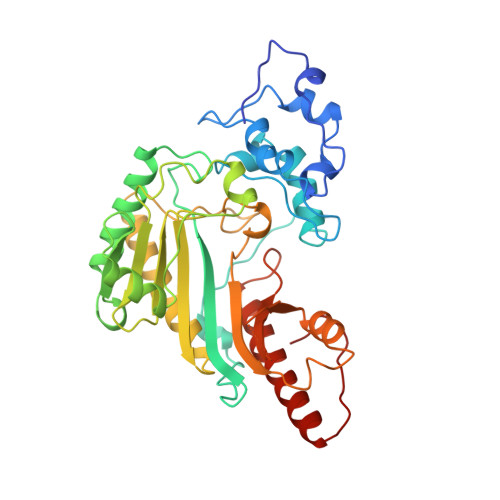
Crystal structure of rabbit muscle creatine kinase.
Rao, J.K., Bujacz, G., Wlodawer, A.(1998) FEBS Lett 439: 133-137
- PubMed: 9849893
- DOI: https://doi.org/10.1016/s0014-5793(98)01355-6
- Primary Citation of Related Structures:
2CRK - PubMed Abstract:
The crystal structure of rabbit muscle creatine kinase, solved at 2.35 A resolution by X-ray diffraction methods, clearly identified the active site with bound sulfates surrounded by a constellation of arginine residues. The putative binding site of creatine, which is occupied by a sulfate group in this analysis, has been tentatively identified. The dimeric interface of the enzyme is held together by a small number of hydrogen bonds.
- Macromolecular Structure Laboratory, NCI-Frederick Cancer Research and Development Center, ABL-Basic Research Program, Frederick, MD 21702, USA. rao@ncifcrf.gov
Organizational Affiliation: