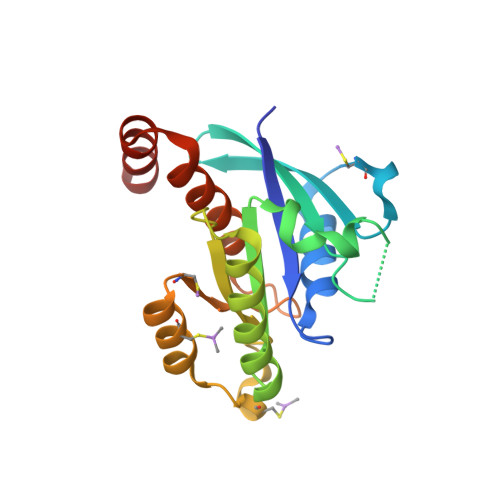
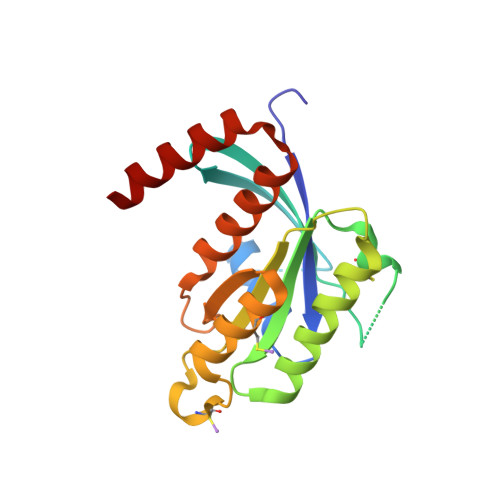
Biochemical and Structural Characterization of the Gem Gtpase.
Splingard, A., Menetrey, J., Perderiset, M., Cicolari, J., Regazzoni, K., Hamoudi, F., Cabanie, L., El Marjou, A., Wells, A., Houdusse, A., De Gunzburg, J.(2007) J Biological Chem 282: 1905
- PubMed: 17107948
- DOI: https://doi.org/10.1074/jbc.M604363200
- Primary Citation of Related Structures:
2CJW - PubMed Abstract:
RGK proteins, encompassing Rad, Gem, Rem1, and Rem2, constitute an intriguing branch of the Ras superfamily; their expression is regulated at the transcription level, they exhibit atypical nucleotide binding motifs, and they carry both large N- and C-terminal extensions. Biochemical and structural studies are required to better understand how such proteins function. Here, we report the first structure for a RGK protein: the crystal structure of a truncated form of the human Gem protein (G domain plus the first part of the C-terminal extension) in complex with Mg.GDP at 2.1 A resolution. It reveals that the G-domain fold and Mg.GDP binding site of Gem are similar to those found for other Ras family GTPases. The first part of the C-terminal extension adopts an alpha-helical conformation that extends along the alpha5 helix and interacts with the tip of the interswitch. Biochemical studies show that the affinities of Gem for GDP and GTP are considerably lower (micromolar range) compared with H-Ras, independent of the presence or absence of N- and C-terminal extensions, whereas its GTPase activity is higher than that of H-Ras and regulated by both extensions. We show how the bulky DXWEX motif, characteristic of the switch II of RGK proteins, affects the conformation of switch I and the phosphate-binding site. Altogether, our data reveal that Gem is a bona fide GTPase that exhibits striking structural and biochemical features that should impact its regulation and cellular activities.
- Institut Curie, Centre de Recherche, Paris F-75248, France, INSERM U528, Paris F-75248, France.
Organizational Affiliation: