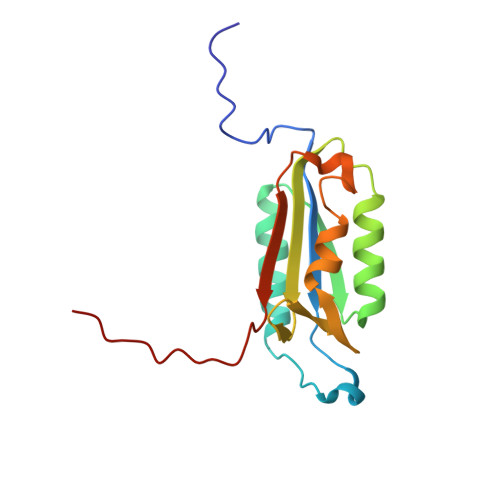
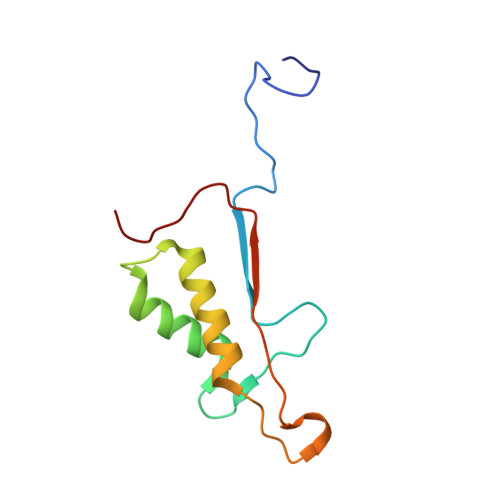
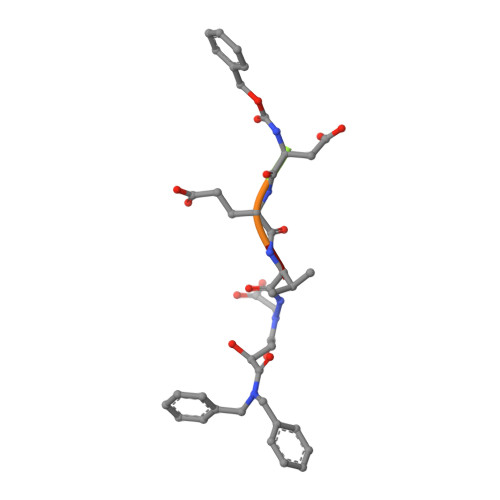
Exploring the S4 and S1 Prime Subsite Specificities in Caspase-3 with Aza-Peptide Epoxide Inhibitors.
Ganesan, R., Jelakovic, S., Campbell, A.J., Li, Z.Z., Asgian, J.L., Powers, J.C., Grutter, M.G.(2006) Biochemistry 45: 9059
- PubMed: 16866351
- DOI: https://doi.org/10.1021/bi060364p
- Primary Citation of Related Structures:
2CDR, 2CNK, 2CNL, 2CNN, 2CNO - PubMed Abstract:
Caspase-3 is a prototypic executioner caspase that plays a central role in apoptosis. Aza-peptide epoxides are a novel class of irreversible inhibitors that are highly specific for clan CD cysteine proteases. The five crystal structures of caspase-3-aza-peptide epoxide inhibitor complexes reported here reveal the structural basis for the mechanism of inhibition and the specificities at the S1' and the S4 subsites. Unlike the clan CA cysteine proteases, the catalytic histidine in caspase-3 plays a critical role during protonation and subsequent ring opening of the epoxide moiety and facilitates the nucleophilic attack by the active site cysteine. The nucleophilic attack takes place on the C3 carbon atom of the epoxide and results in an irreversible alkylation of the active site cysteine residue. A favorable network of hydrogen bonds involving the oxyanion hole, catalytic histidine, and the atoms in the prime site of the inhibitor enhance the binding affinity and specificity of the aza-peptide epoxide inhibitors toward caspase-3. The studies also reveal that subtle movements of the N-terminal loop of the beta-subunit occur when the P4 Asp is replaced by a P4 Ile, whereas the N-terminal loop and the safety catch Asp179 are completely disordered when the P4 Asp is replaced by P4 Cbz group.
- Department of Biochemistry, University of Zurich, Winterthurerstrasse 190, CH-8057 Zurich, Switzerland.
Organizational Affiliation: