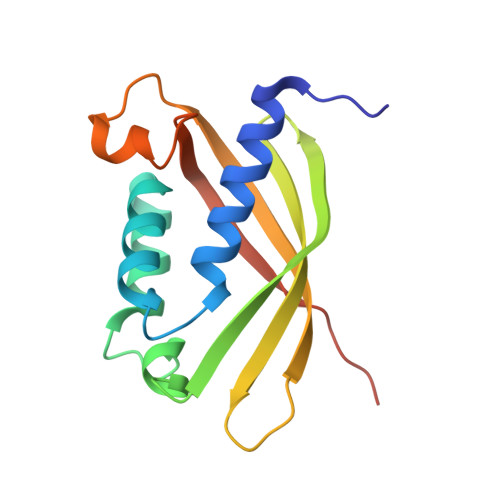
Agrobacterium Tumefaciens Virb8 Structure Reveals Potential Protein-Protein Interactions Sites.
Bailey, S., Ward, D., Middleton, R., Grossmann, J.G., Zambryski, P.C.(2006) Proc Natl Acad Sci U S A 103: 2582
- PubMed: 16481621
- DOI: https://doi.org/10.1073/pnas.0511216103
- Primary Citation of Related Structures:
2CC3 - PubMed Abstract:
Bacterial type IV secretion systems (T4SS) translocate DNA and/or proteins to recipient cells, thus providing a mechanism for conjugative transfer of genetic material and bacterial pathogenesis. Here we describe the first structure of a core component from the archetypal Agrobacterium tumefaciens T4SS: the 2.2-A resolution crystal structure of the VirB8 periplasmic domain (pVirB8(AT)). VirB8 forms a dimer in the crystal, and we identify residues likely important for stabilization of the dimer interface. Structural comparison of pVirB8(AT) with Brucella suis VirB8 confirms that the monomers have a similar fold. In addition, the pVirB8(AT) dimer superimposes very closely on the B. suis VirB8 dimer, supporting the proposal that dimer formation in the crystal reflects self-interactions that are biologically significant. The evolutionary conservation level for each residue was obtained from a data set of 84 VirB8 homologs and projected onto the protein structure to indicate conserved surface patches that likely contact other T4SS proteins.
- Molecular Biophysics Group, Council for the Central Laboratory of the Research Councils Daresbury Laboratory, Warrington WA4 4AD, United Kingdom.
Organizational Affiliation: