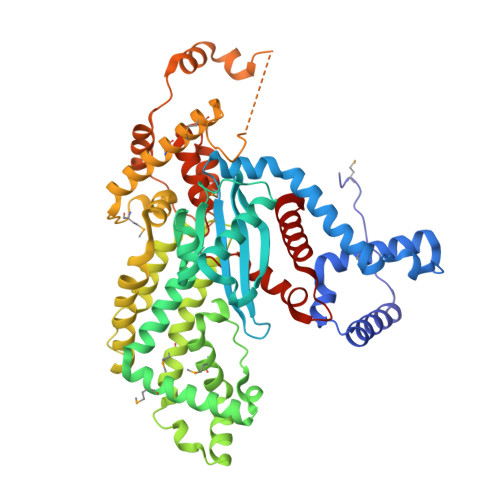
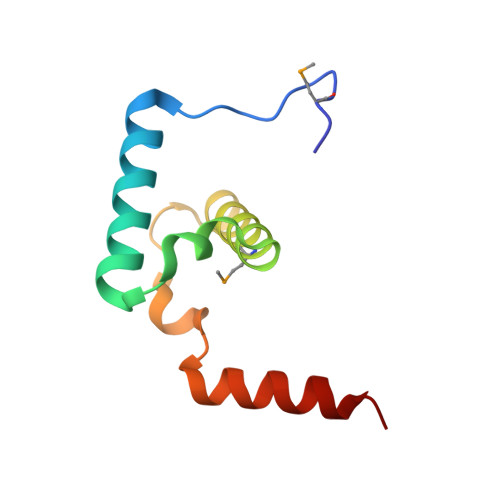
Molecular Architecture of a Eukaryotic DNA Transposase
Hickman, A.B., Perez, Z.N., Zhou, L., Musingarimi, P., Ghirlando, R., Hinshaw, J.E., Craig, N.L., Dyda, F.(2005) Nat Struct Mol Biol 12: 715
- PubMed: 16041385
- DOI: https://doi.org/10.1038/nsmb970
- Primary Citation of Related Structures:
2BW3 - PubMed Abstract:
Mobile elements and their inactive remnants account for large proportions of most eukaryotic genomes, where they have had central roles in genome evolution. Over 50 years ago, McClintock reported a form of stress-induced genome instability in maize in which discrete DNA segments move between chromosomal locations. Our current mechanistic understanding of enzymes catalyzing transposition is largely limited to prokaryotic transposases. The Hermes transposon from the housefly is part of the eukaryotic hAT superfamily that includes hobo from Drosophila, McClintock's maize Activator and Tam3 from snapdragon. We report here the three-dimensional structure of a functionally active form of the transposase from Hermes at 2.1-A resolution. The Hermes protein has some structural features of prokaryotic transposases, including a domain with a retroviral integrase fold. However, this domain is disrupted by the insertion of an additional domain. Finally, transposition is observed only when Hermes assembles into a hexamer.
- Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, Maryland 20892, USA.
Organizational Affiliation: