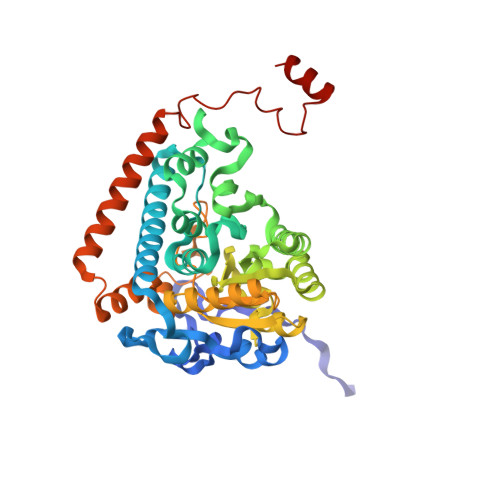
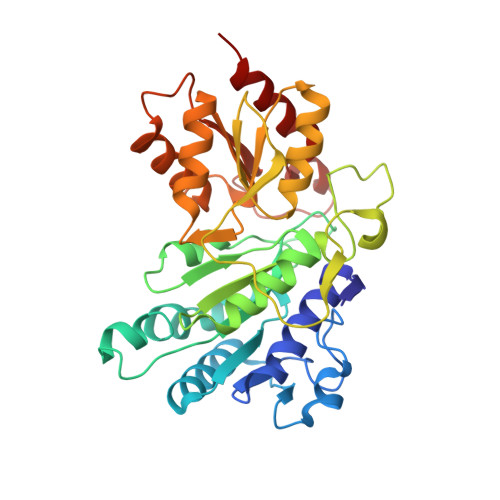
The Molecular Origins of Specificity in the Assembly of a Multienzyme Complex.
Frank, R.A.W., Pratap, J.V., Pei, X.Y., Perham, R.N., Luisi, B.F.(2005) Structure 13: 1119
- PubMed: 16084384
- DOI: https://doi.org/10.1016/j.str.2005.04.021
- Primary Citation of Related Structures:
2BP7 - PubMed Abstract:
The pyruvate dehydrogenase (PDH) multienzyme complex is central to oxidative metabolism. We present the first crystal structure of a complex between pyruvate decarboxylase (E1) and the peripheral subunit binding domain (PSBD) of the dihydrolipoyl acetyltransferase (E2). The interface is dominated by a "charge zipper" of networked salt bridges. Remarkably, the PSBD uses essentially the same zipper to alternately recognize the dihydrolipoyl dehydrogenase (E3) component of the PDH assembly. The PSBD achieves this dual recognition largely through the addition of a network of interfacial water molecules unique to the E1-PSBD complex. These structural comparisons illuminate our observations that the formation of this water-rich E1-E2 interface is largely enthalpy driven, whereas that of the E3-PSBD complex (from which water is excluded) is entropy driven. Interfacial water molecules thus diversify surface complementarity and contribute to avidity, enthalpically. Additionally, the E1-PSBD structure provides insight into the organization and active site coupling within the approximately 9 MDa PDH complex.
- Department of Biochemistry, University of Cambridge, 80 Tennis Court Road, Cambridge CB2 1GA, United Kingdom.
Organizational Affiliation: