Structural basis for transcription inhibition by tagetitoxin
Vassylyev, D.G., Svetlov, V., Vassylyeva, M.N., Perederina, A., Igarashi, N., Matsugaki, N., Wakatsuki, S., Artsimovitch, I.(2005) Nat Struct Mol Biol 12: 1086-1093
- PubMed: 16273103
- DOI: https://doi.org/10.1038/nsmb1015
- Primary Citation of Related Structures:
2BE5 - PubMed Abstract:
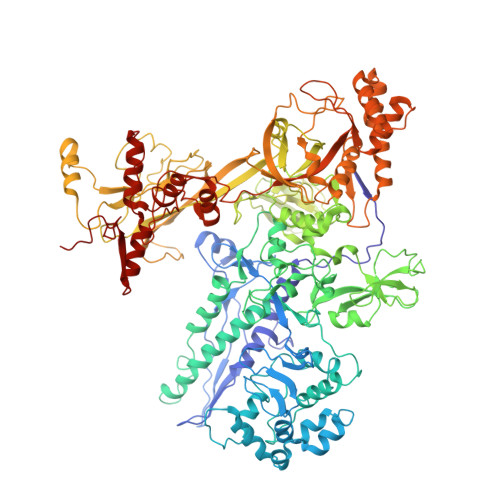
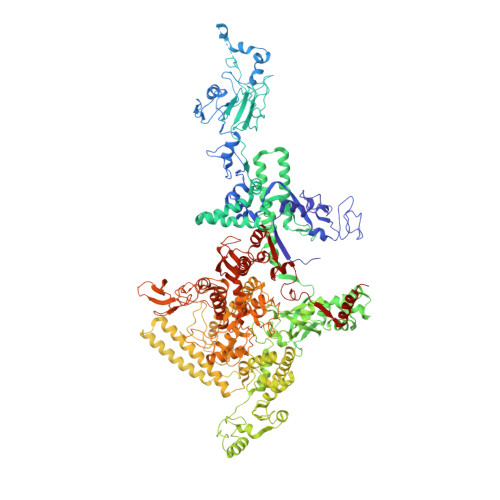
Tagetitoxin (Tgt) inhibits transcription by an unknown mechanism. A structure at a resolution of 2.4 A of the Thermus thermophilus RNA polymerase (RNAP)-Tgt complex revealed that the Tgt-binding site within the RNAP secondary channel overlaps that of the stringent control effector ppGpp, which partially protects RNAP from Tgt inhibition. Tgt binding is mediated exclusively through polar interactions with the beta and beta' residues whose substitutions confer resistance to Tgt in vitro. Importantly, a Tgt phosphate, together with two active site acidic residues, coordinates the third Mg(2+) ion, which is distinct from the two catalytic metal ions. We show that Tgt inhibits all RNAP catalytic reactions and propose a mechanism in which the Tgt-bound Mg(2+) ion has a key role in stabilization of an inactive transcription intermediate. Remodeling of the active site by metal ions could be a common theme in the regulation of catalysis by nucleic acid enzymes.
- Department of Biochemistry and Molecular Genetics, University of Alabama at Birmingham, School of Medicine, Birmingham, Alabama 35294, USA. dmitry@uab.edu
Organizational Affiliation: