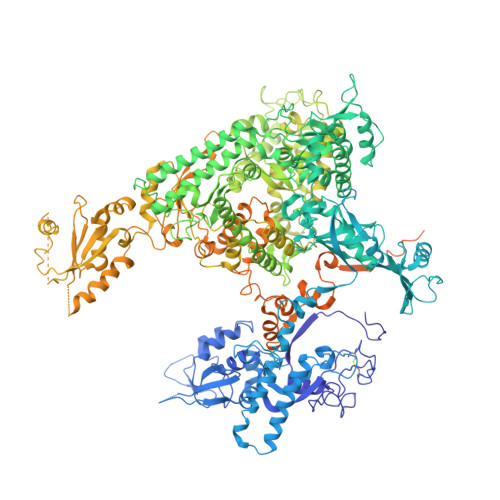
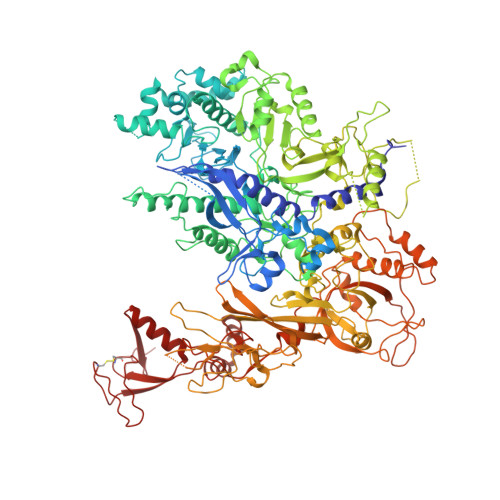
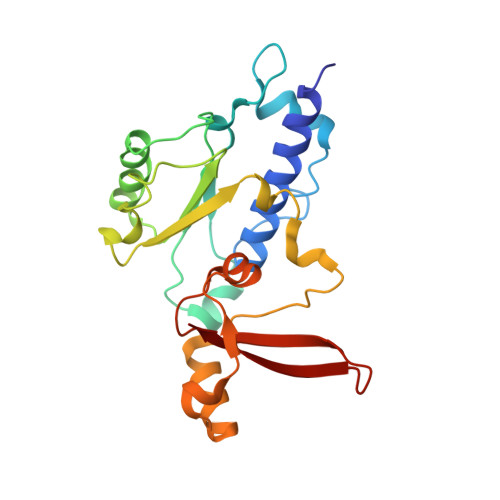
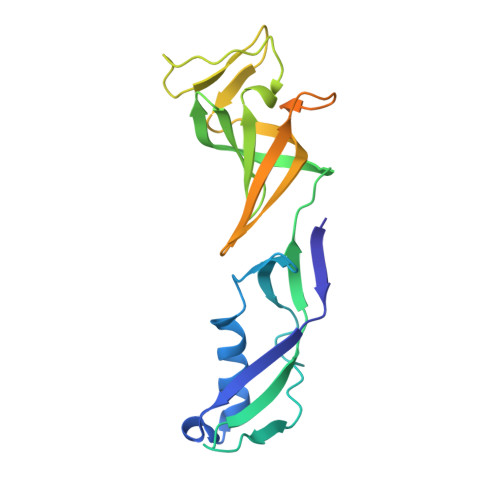
Phasing RNA Polymerase II Using Intrinsically Bound Zn Atoms: An Updated Structural Model.
Meyer, P.A., Ye, P., Zhang, M., Suh, M.H., Fu, J.(2006) Structure 14: 973-982
- PubMed: 16765890
- DOI: https://doi.org/10.1016/j.str.2006.04.003
- Primary Citation of Related Structures:
2B8K - PubMed Abstract:
Macromolecular assemblies as large as RNA polymerase II (Pol II) can be phased by a few intrinsically bound Zn atoms, by using MAD experiments as described here. A phasing effectiveness of 570 aa/Zn is attained for Pol II. The resulting experimental, unbiased electron density map is of such quality that it confirms the existing crystallographic model and further reveals structural regions not shown by model phases, thus updating the Pol II model at three sites. The mechanistically important fork loop-1 element is observed to be ordered in the absence of nucleic acids, suggesting additional insights into the mechanisms that maintain the stability of the transcription ternary complex and allow its release. Furthermore, a computational experiment with simulated MAD data sets demonstrates that 1 Zn site is able to provide adequate experimental phase information for as many as 1100 amino acids of polypeptide, under the conditions of the current synchrotron and detector technologies.
- Department of Molecular Biology and Genetics, Cornell University, 221 Biotechnology Building, Ithaca, New York 14853, USA.
Organizational Affiliation: