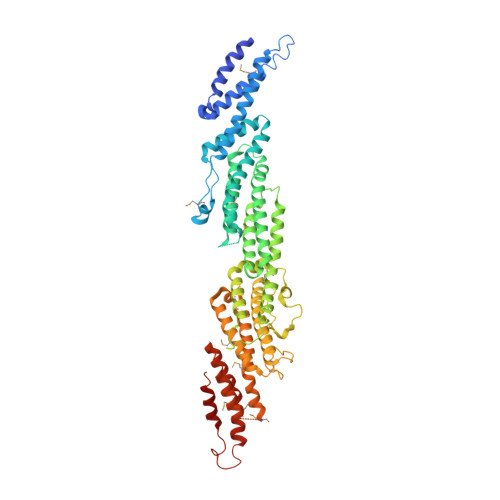
Crystal Structure of the S.cerevisiae Exocyst Component Exo70p
Hamburger, Z.A., Hamburger, A.E., West, A.P., Weis, W.I.(2006) J Mol Biology 356: 9-21
- PubMed: 16359701
- DOI: https://doi.org/10.1016/j.jmb.2005.09.099
- Primary Citation of Related Structures:
2B7M - PubMed Abstract:
The exocyst is an evolutionarily conserved multiprotein complex required for the targeting and docking of post-Golgi vesicles to the plasma membrane. Through its interactions with a variety of proteins, including small GTPases, the exocyst is thought to integrate signals from the cell and signal that vesicles arriving at the plasma membrane are ready for fusion. Here we describe the three-dimensional crystal structure of one of the components of the exocyst, Exo70p, from Saccharomyces cerevisiae at 3.5A resolution. Exo70p binds the small GTPase Rho3p in a GTP-dependent manner with an equilibrium dissociation constant of approximately 70 microM. Exo70p is an extended rod approximately 155 angstroms in length composed principally of alpha helices, and is a novel fold. The structure provides a first view of the Exo70 protein family and provides a framework to study the molecular function of this exocyst component.
- Department of Structural Biology, Stanford University School of Medicine, Stanford, CA 94305-5126, USA.
Organizational Affiliation: